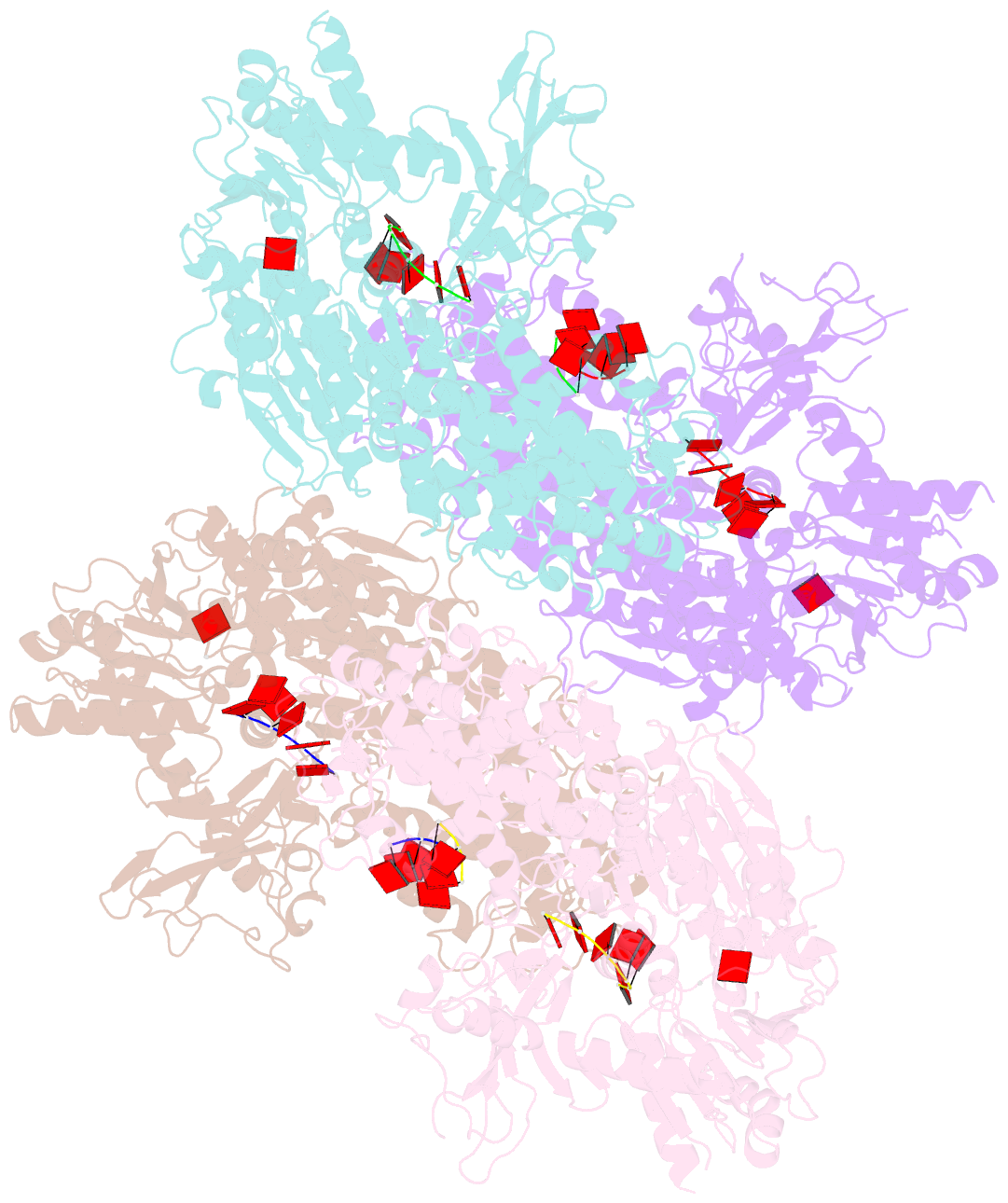
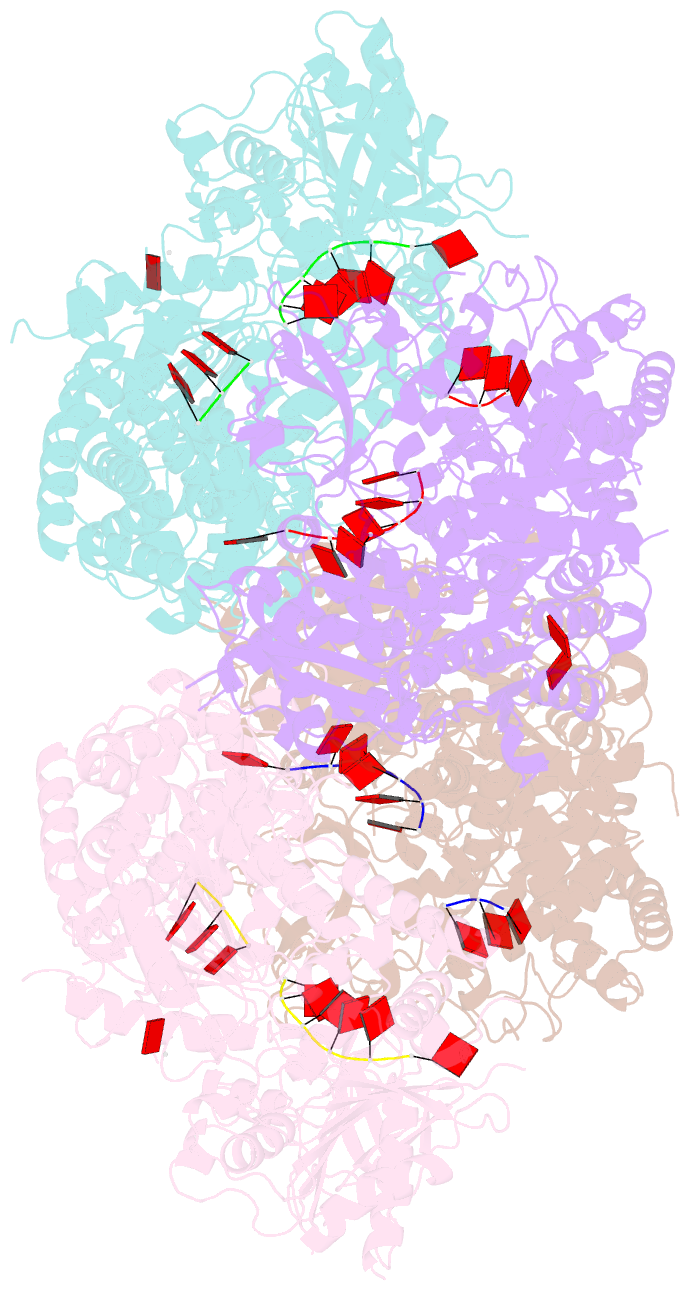
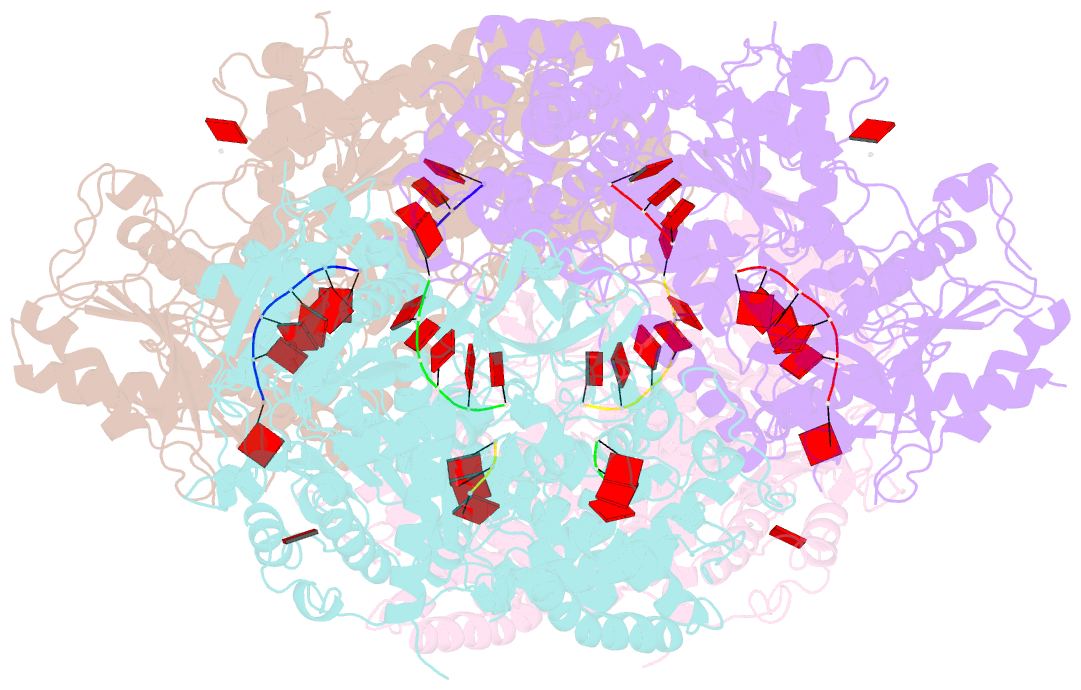
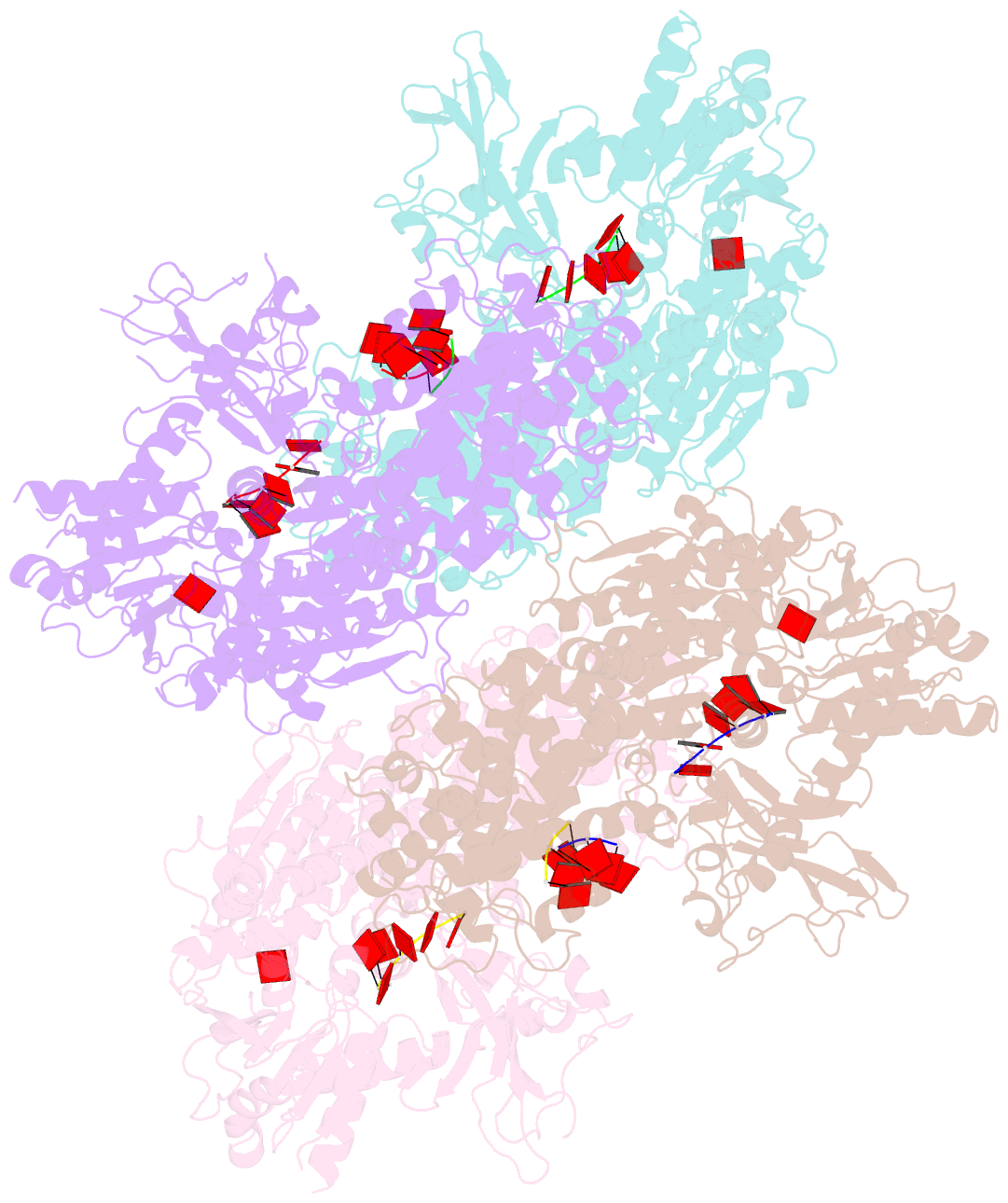
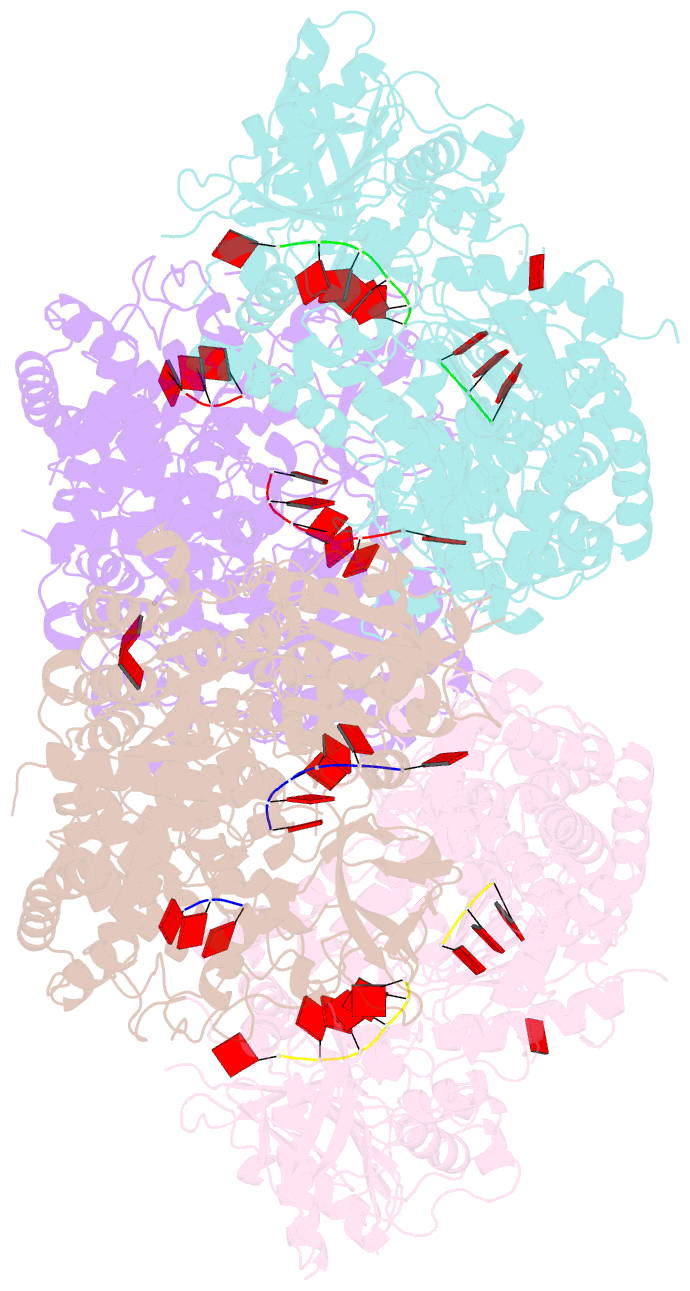
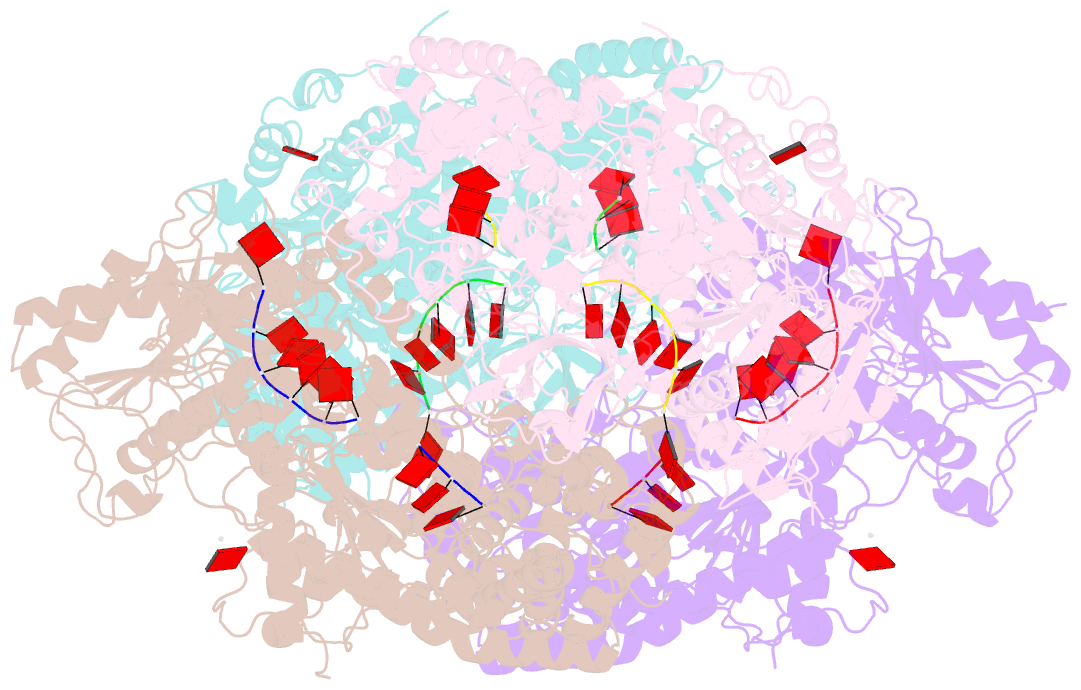
Summary information and primary citation
- PDB-id
- 4qqy; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (3.12 Å)
- Summary
- Crystal structure of t. fusca cas3-adp
- Reference
- Huo Y, Nam KH, Ding F, Lee H, Wu L, Xiao Y, Farchione MD, Zhou S, Rajashankar K, Kurinov I, Zhang R, Ke A (2014): "Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation." Nat.Struct.Mol.Biol., 21, 771-777. doi: 10.1038/nsmb.2875.
- Abstract
- CRISPR drives prokaryotic adaptation to invasive nucleic acids such as phages and plasmids, using an RNA-mediated interference mechanism. Interference in type I CRISPR-Cas systems requires a targeting Cascade complex and a degradation machine, Cas3, which contains both nuclease and helicase activities. Here we report the crystal structures of Thermobifida fusca Cas3 bound to single-stranded (ss) DNA substrate and show that it is an obligate 3'-to-5' ssDNase that preferentially accepts substrate directly from the helicase moiety. Conserved residues in the HD-type nuclease coordinate two irons for ssDNA cleavage. We demonstrate ATP coordination and conformational flexibility of the SF2-type helicase domain. Cas3 is specifically guided toward Cascade-bound target DNA by a PAM sequence, through physical interactions with both the nontarget substrate strand and the CasA protein. The sequence of recognition events ensures well-controlled DNA targeting and degradation of foreign DNA by Cascade and Cas3.