Summary information and primary citation
- PDB-id
- 4qvd; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (1.972 Å)
- Summary
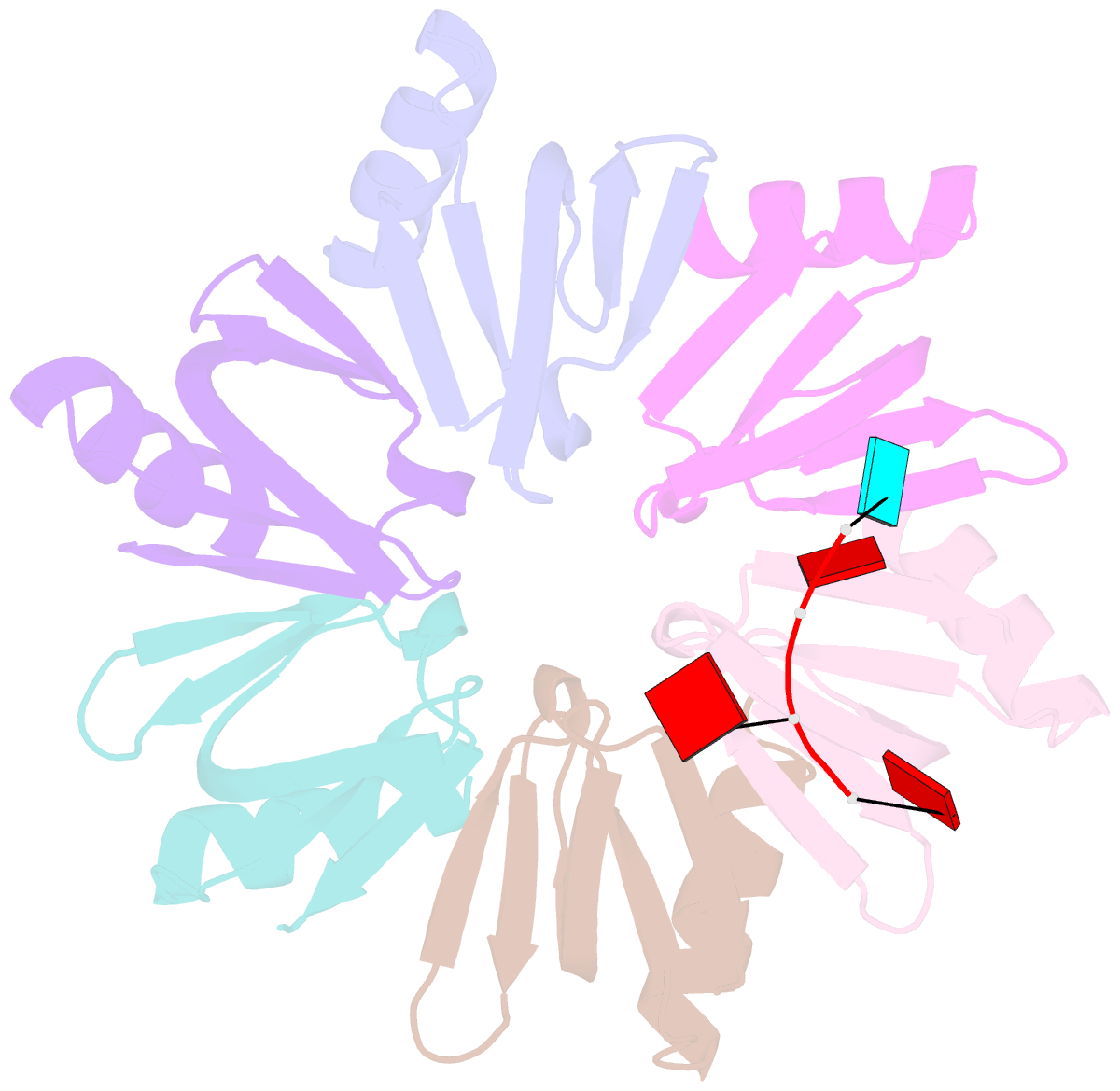
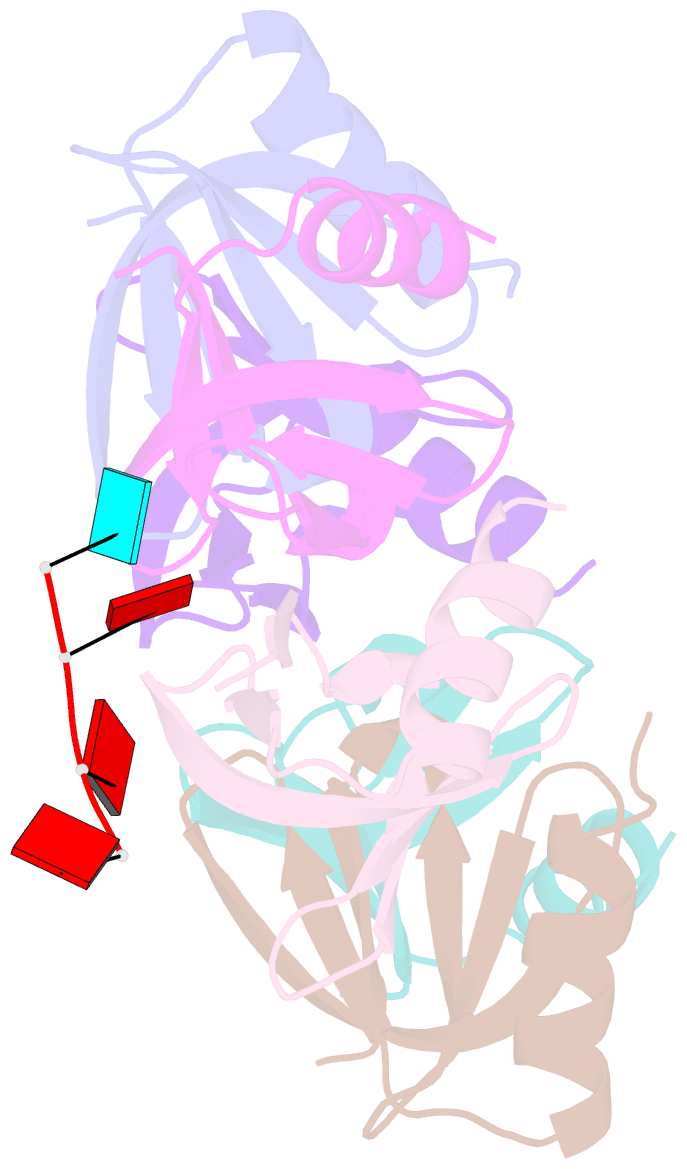
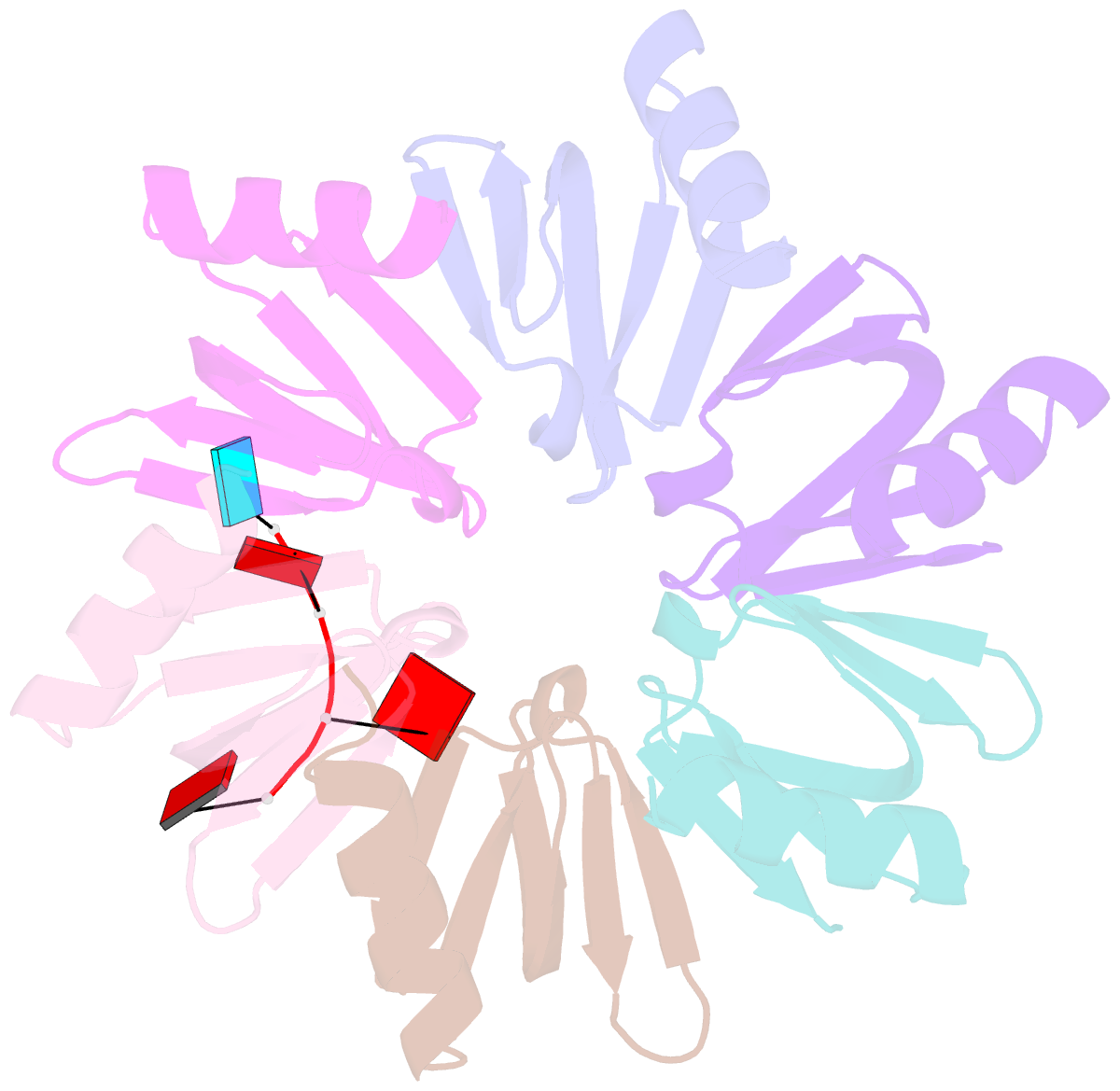
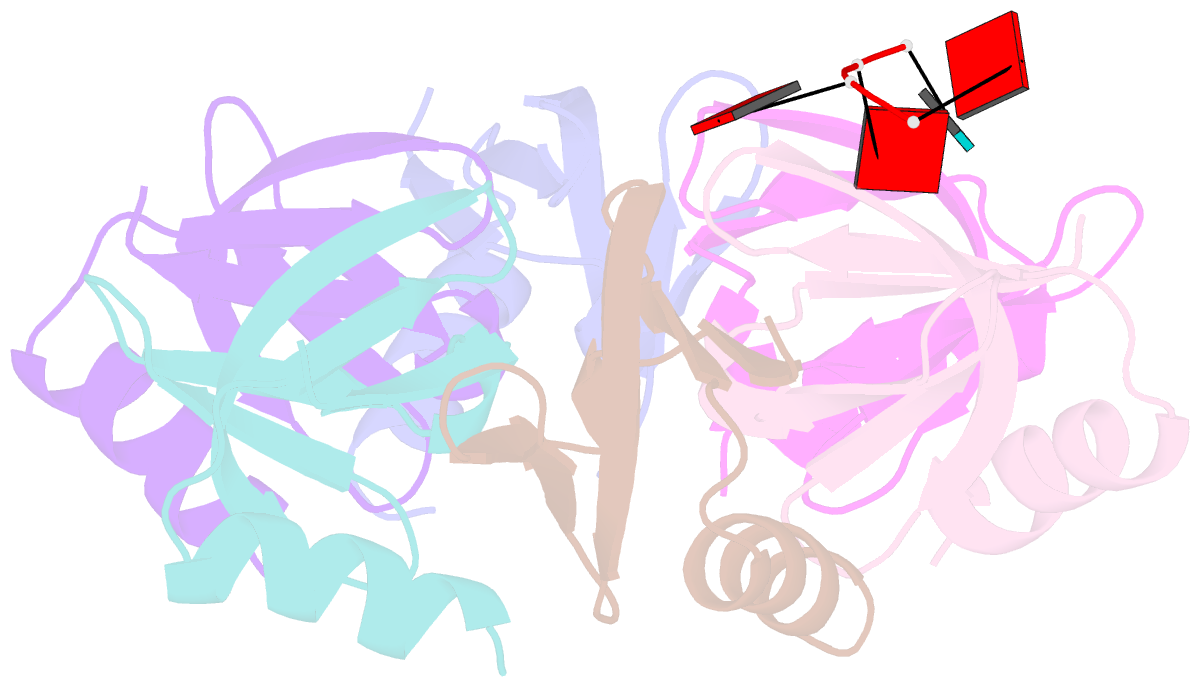
- E.coli hfq in complex with RNA ads
- Reference
- Wang LJ, Wang WW, Li FD, Zhang J, Wu JH, Gong QG, Shi YY (2015): "Structural insights into the recognition of the internal A-rich linker from OxyS sRNA by Escherichia coli Hfq." Nucleic Acids Res., 43, 2400-2411. doi: 10.1093/nar/gkv072.
- Abstract
- Small RNA OxyS is induced during oxidative stress in Escherichia coli and it is an Hfq-dependent negative regulator of mRNA translation. OxyS represses the translation of fhlA and rpoS mRNA, which encode the transcriptional activator and σ(s) subunit of RNA polymerase, respectively. However, little is known regarding how Hfq, an RNA chaperone, interacts with OxyS at the atomic level. Here, using fluorescence polarization and tryptophan fluorescence quenching assays, we verified that the A-rich linker region of OxyS sRNA binds Hfq at its distal side. We also report two crystal structures of Hfq in complex with A-rich RNA fragments from this linker region. Both of these RNA fragments bind to the distal side of Hfq and adopt a different conformation compared with those previously reported for the (A-R-N)n tripartite recognition motif. Furthermore, using fluorescence polarization, electrophoresis mobility shift assays and in vivo translation assays, we found that an Hfq mutant, N48A, increases the binding affinity of OxyS for Hfq in vitro but is defective in the negative regulation of fhlA translation in vivo, suggesting that the normal function of OxyS depends on the details of the interaction with Hfq that may be related to the rapid recycling of Hfq in the cell.