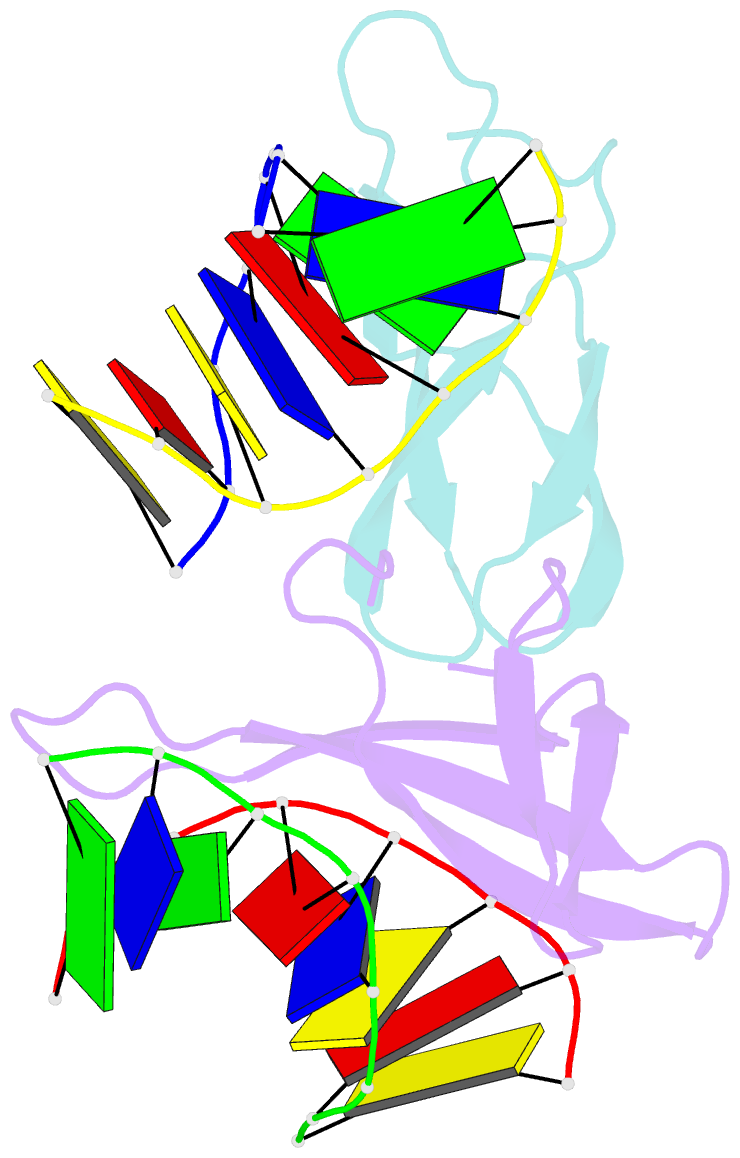
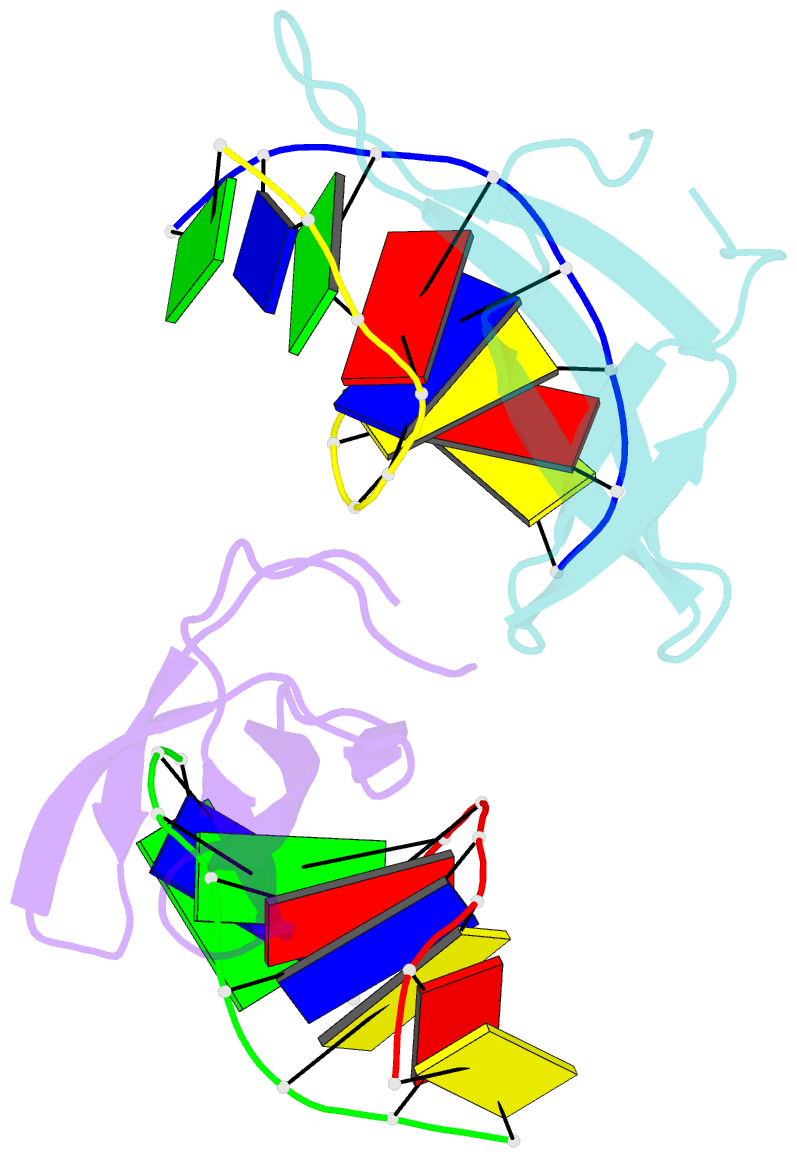
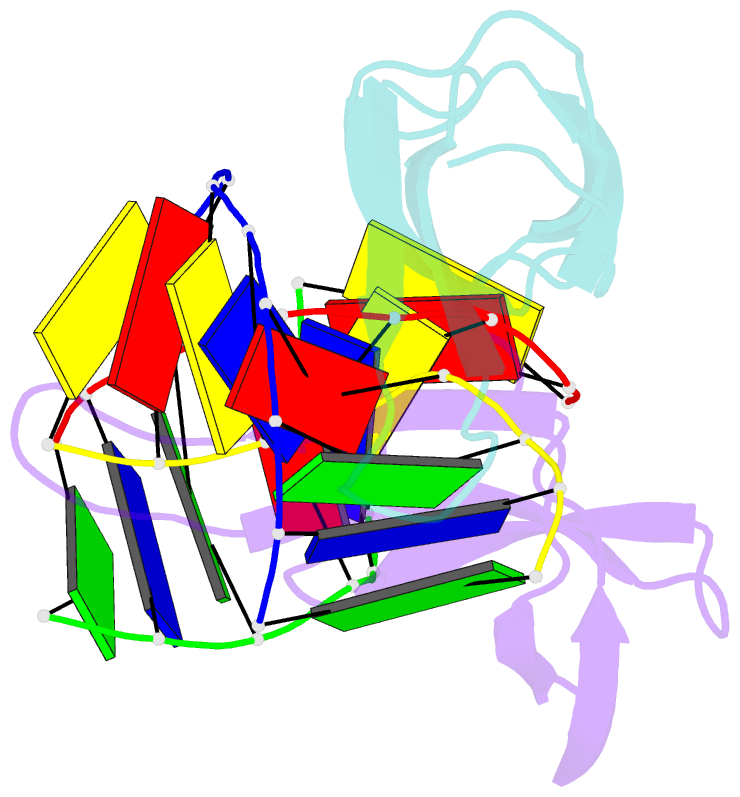
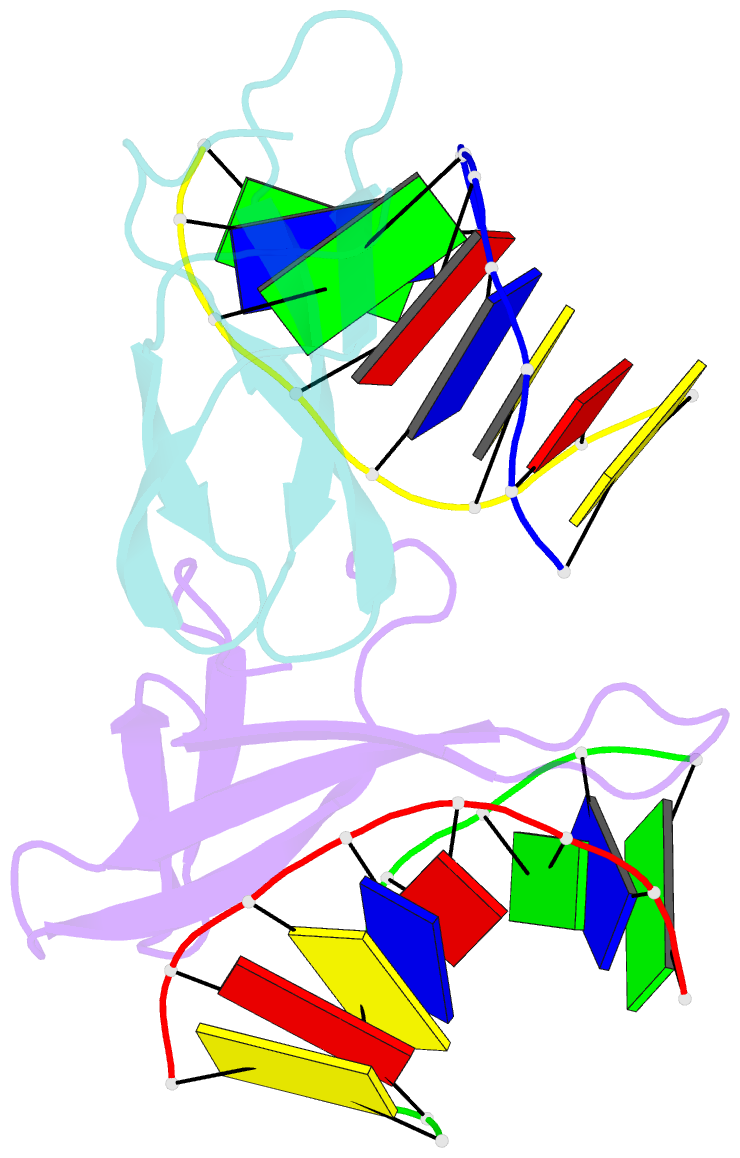
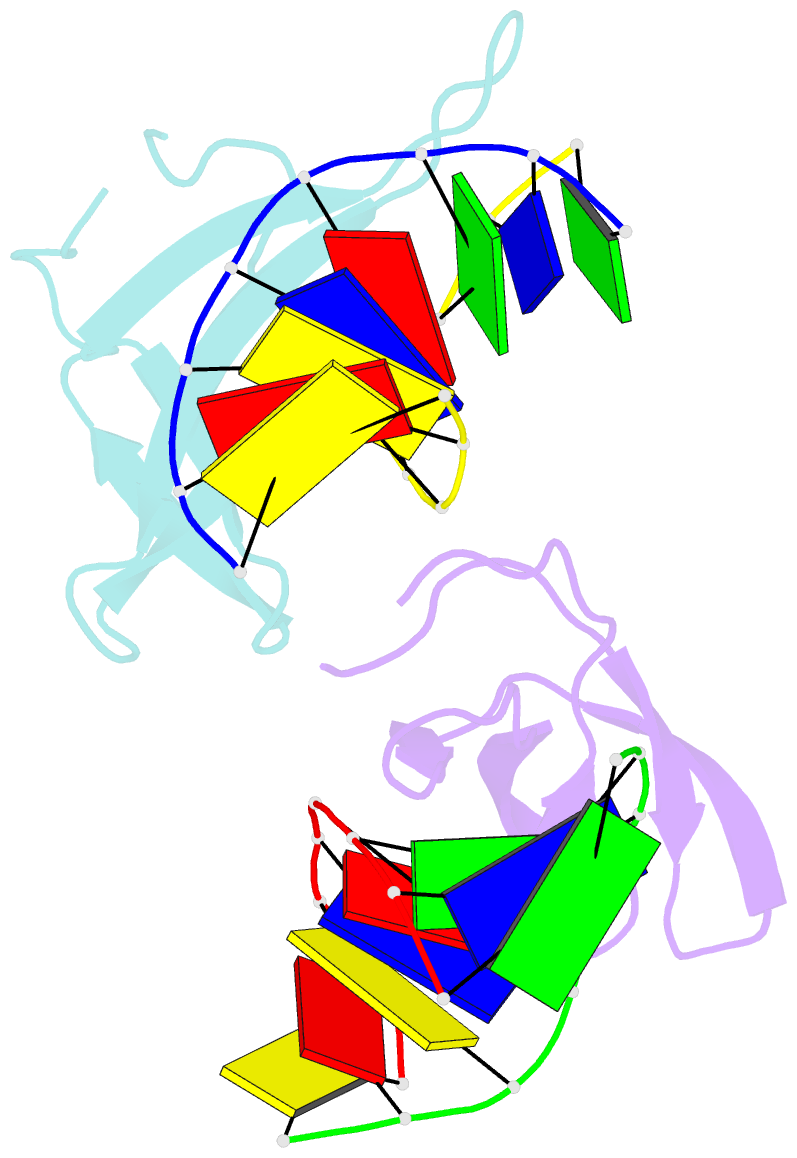
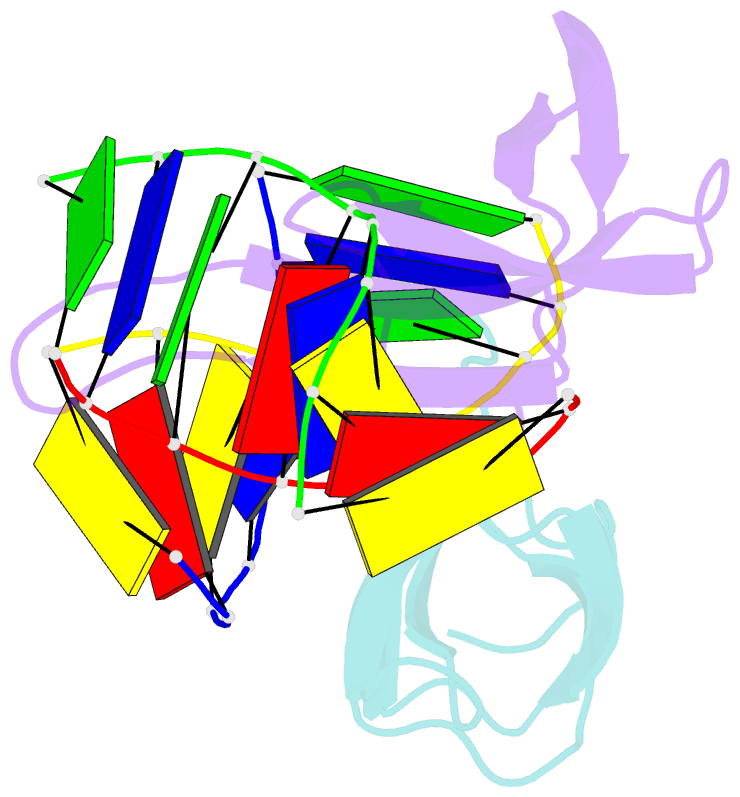
Summary information and primary citation
- PDB-id
- 4r56; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.3 Å)
- Summary
- Crystal structure of sulfolobus cren7-dsDNA(gtgatcac) complex
- Reference
- Zhang ZF, Gong Y, Chen YY, Li HB, Huang L (2015): "Insights into the interaction between Cren7 and DNA: the role of loop beta 3-beta 4." Extremophiles, 19, 395-406. doi: 10.1007/s00792-014-0725-y.
- Abstract
- Sulfolobus synthesizes large amounts of small chromatin proteins Cren7 and Sul7d. The two proteins share overall structural similarity, but differ distinctly in the DNA-binding region between β3- and β4-strands. While Sul7d possesses a hinge of two amino acid residues, Cren7 contains a flexible seven-residue loop (loop β3-β4) in the region. Here, we report the role of loop β3-β4 in the interaction of Cren7 with duplex DNA. We show that all residues with a large side chain on the loop, i.e., Pro30, Lys31, Arg33 and Lys34, contributed significantly to the binding of Cren7 to DNA. The three basic amino acids affected the ability of Cren7 to constrain negative DNA supercoils in a residue number-dependent manner. The crystal structure of a complex between a mutant Cren7 protein (GR) with loop β3-β4 replaced by two residues (Gly and Arg) to mimic the hinge at the corresponding position in Sul7d and an 8-bp dsDNA has been determined. Structural comparison between the GR-DNA and Cren7-DNA complexes shows that GR resembles Sul7d more than Cren7 in DNA-binding size and in the effect on the width of the major groove of DNA and the pattern of DNA bending. However, GR induces smaller DNA curvature than Sul7d. Our results suggest that Cren7 and Sul7d package chromosomal DNA in a slightly different fashion, presumably permitting different chromosomal accessibility by proteins functioning in DNA transactions.