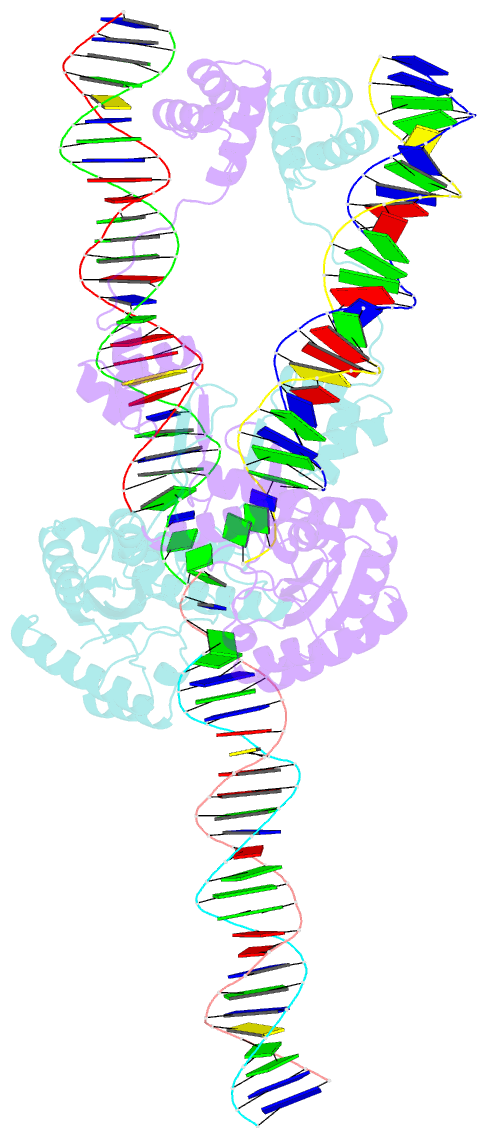
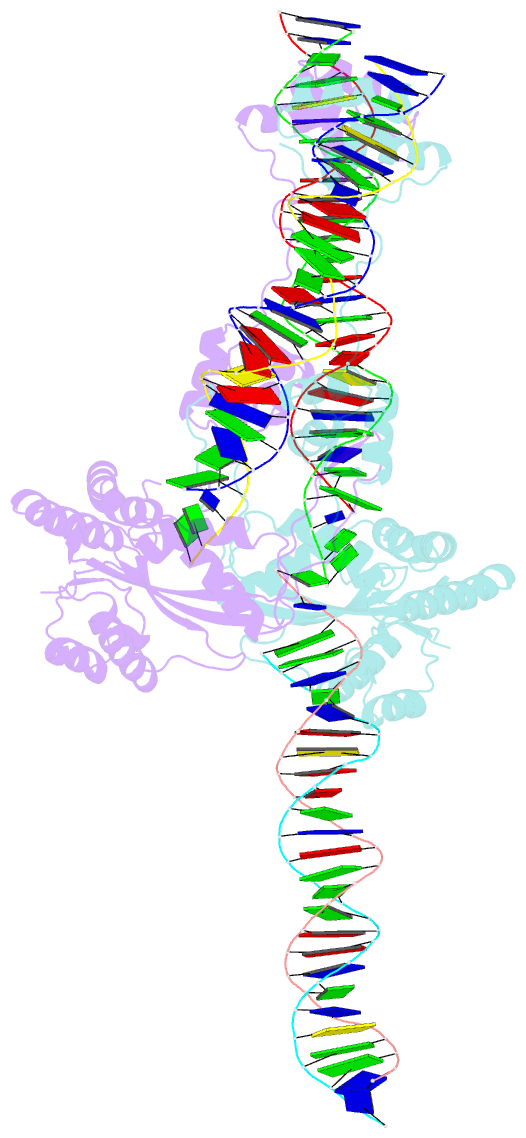
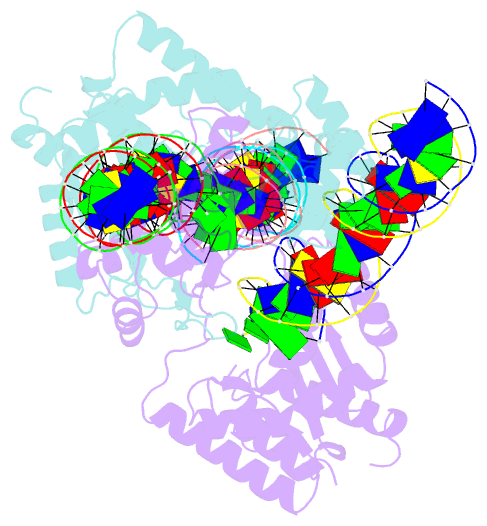
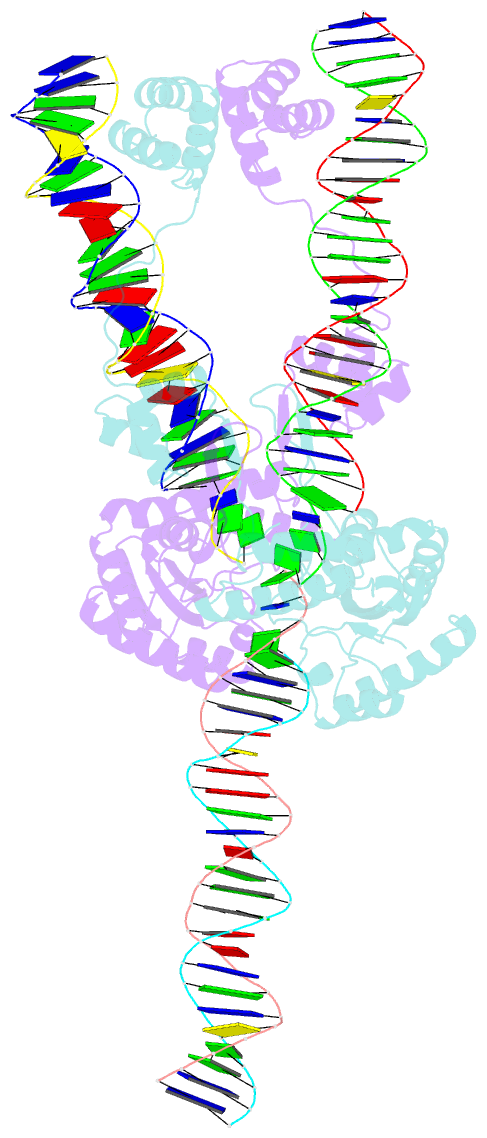
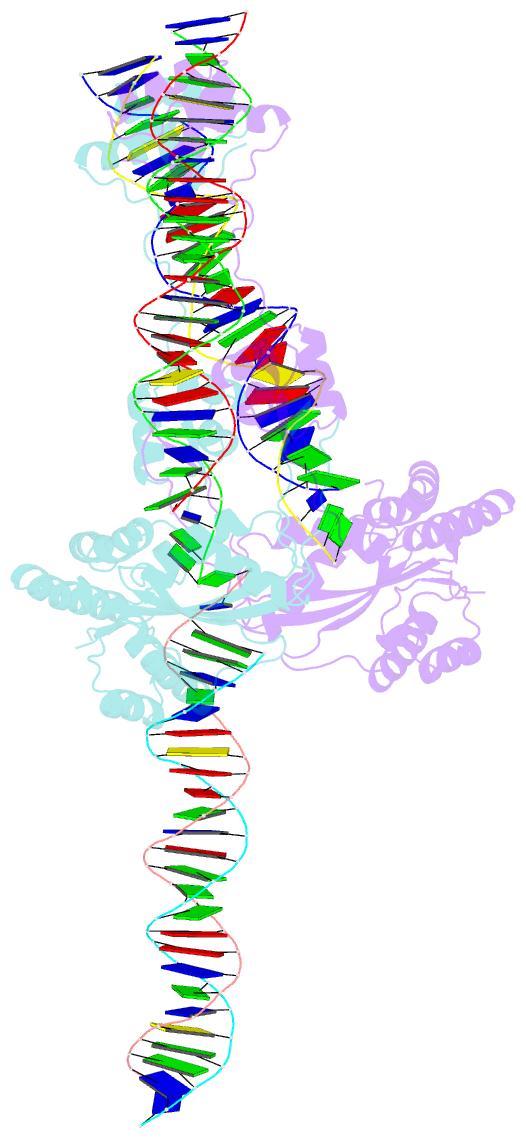
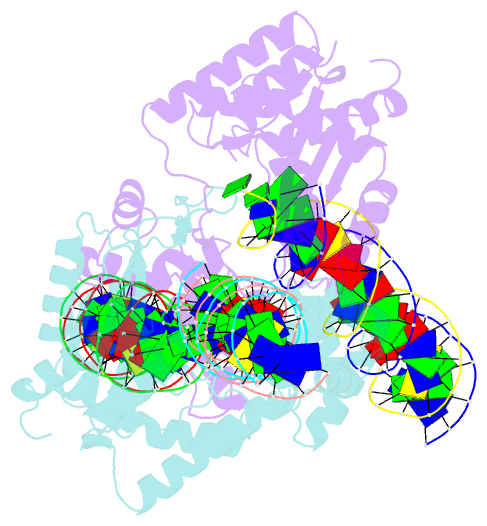
Summary information and primary citation
- PDB-id
- 4r79; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- recombination-DNA
- Method
- X-ray (3.1 Å)
- Summary
- Mos1 transposase paired-end complex with left transposon end
- Reference
- Trubitsyna M, Grey H, Houston DR, Finnegan DJ, Richardson JM (2015): "Structural Basis for the Inverted Repeat Preferences of mariner Transposases." J.Biol.Chem., 290, 13531-13540. doi: 10.1074/jbc.M115.636704.
- Abstract
- The inverted repeat (IR) sequences delimiting the left and right ends of many naturally active mariner DNA transposons are non-identical and have different affinities for their transposase. We have compared the preferences of two active mariner transposases, Mos1 and Mboumar-9, for their imperfect transposon IRs in each step of transposition: DNA binding, DNA cleavage, and DNA strand transfer. A 3.1 Å resolution crystal structure of the Mos1 paired-end complex containing the pre-cleaved left IR sequences reveals the molecular basis for the reduced affinity of the Mos1 transposase DNA-binding domain for the left IR as compared with the right IR. For both Mos1 and Mboumar-9, in vitro DNA transposition is most efficient when the preferred IR sequence is present at both transposon ends. We find that this is due to the higher efficiency of cleavage and strand transfer of the preferred transposon end. We show that the efficiency of Mboumar-9 transposition is improved almost 4-fold by changing the 3' base of the preferred Mboumar-9 IR from guanine to adenine. This preference for adenine at the reactive 3' end for both Mos1 and Mboumar-9 may be a general feature of mariner transposition.