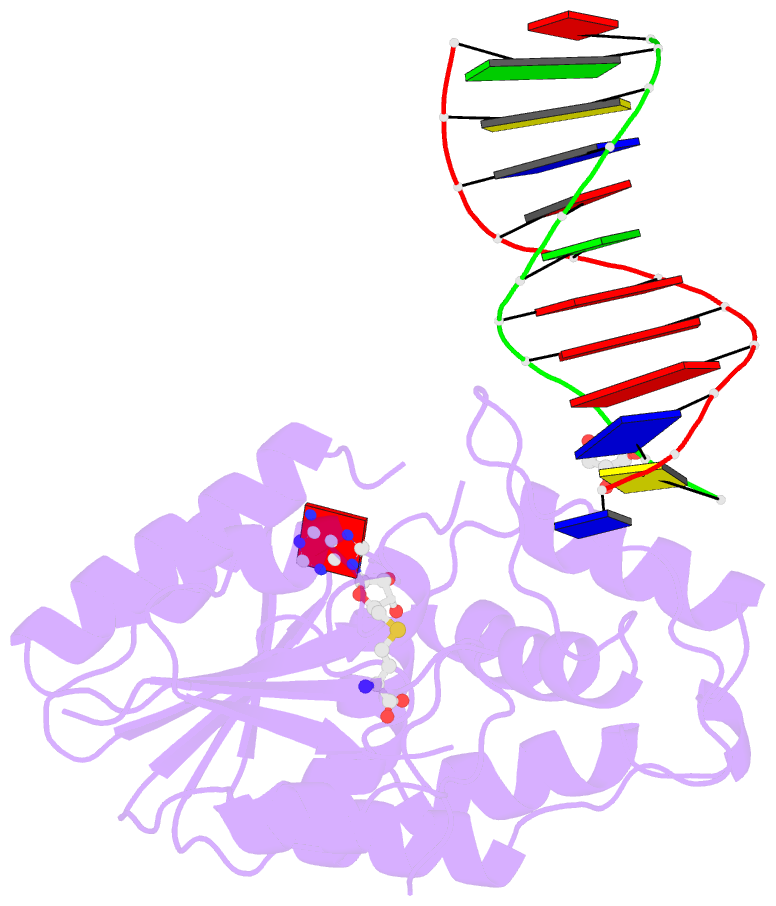
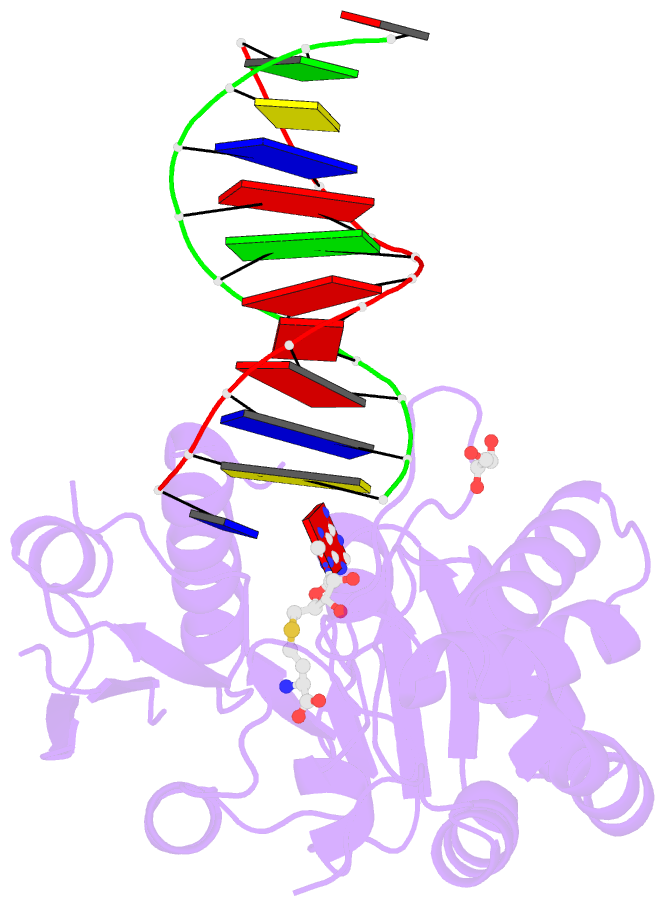
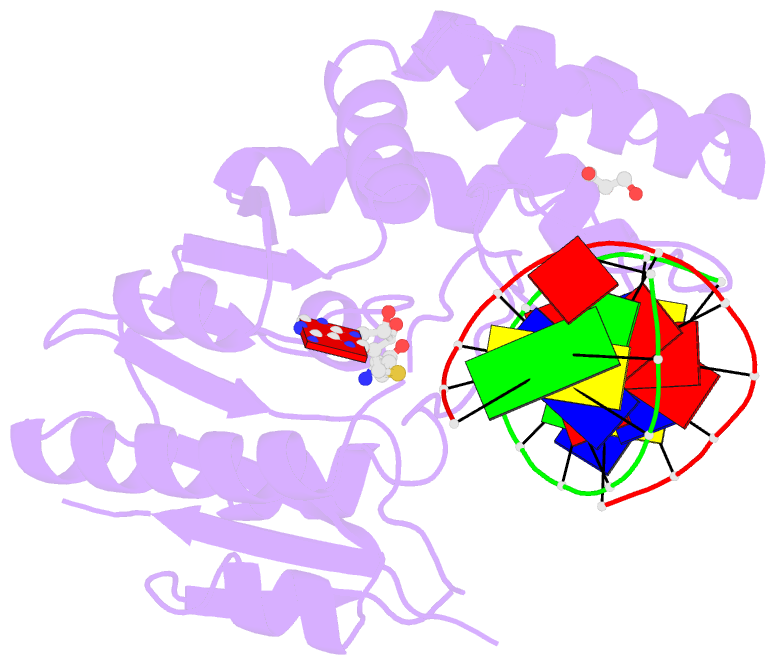
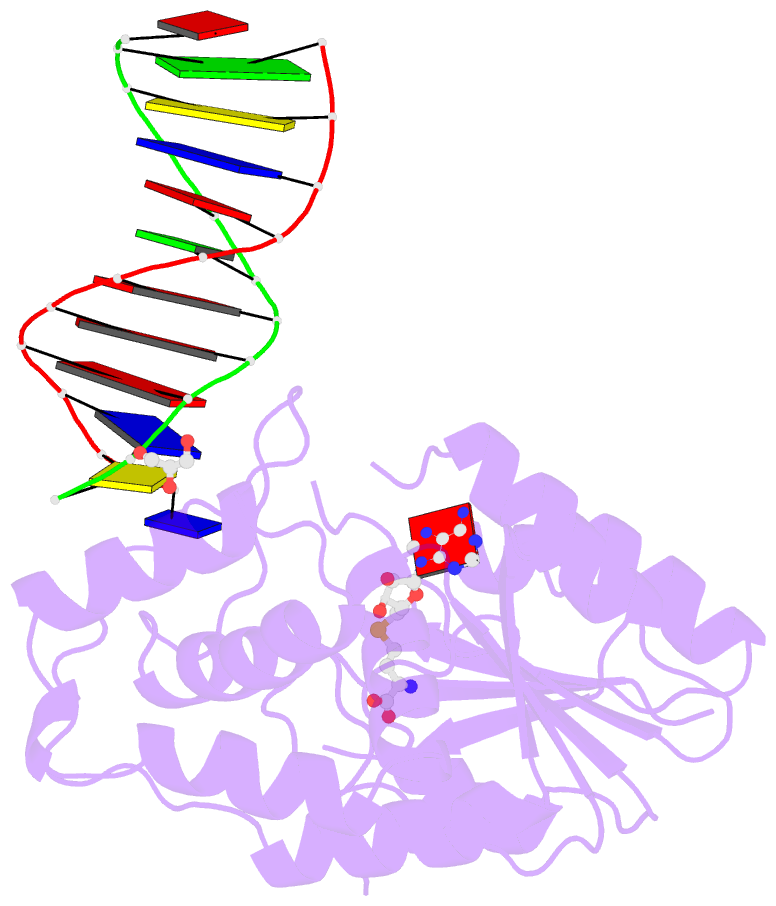
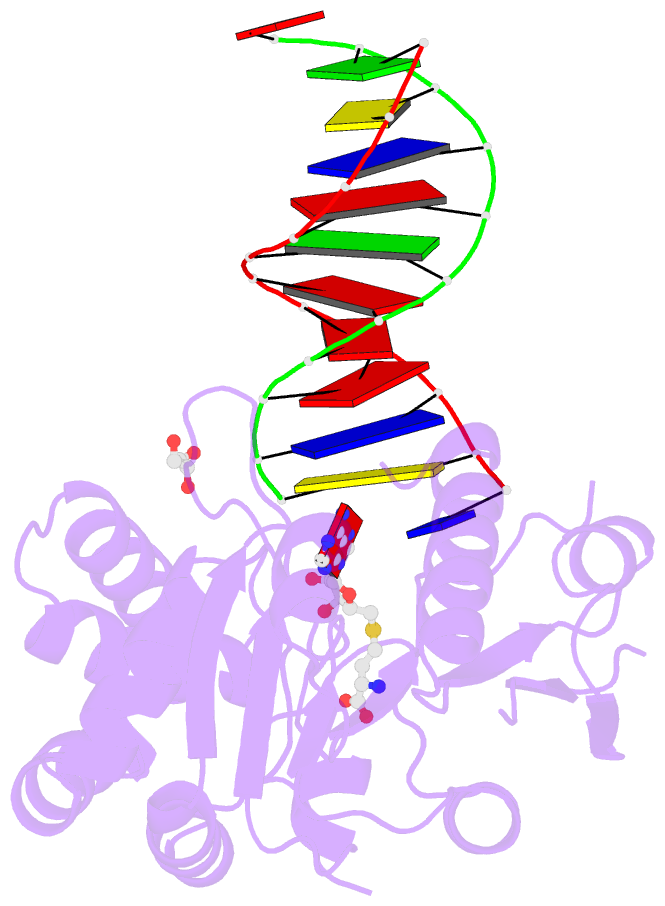
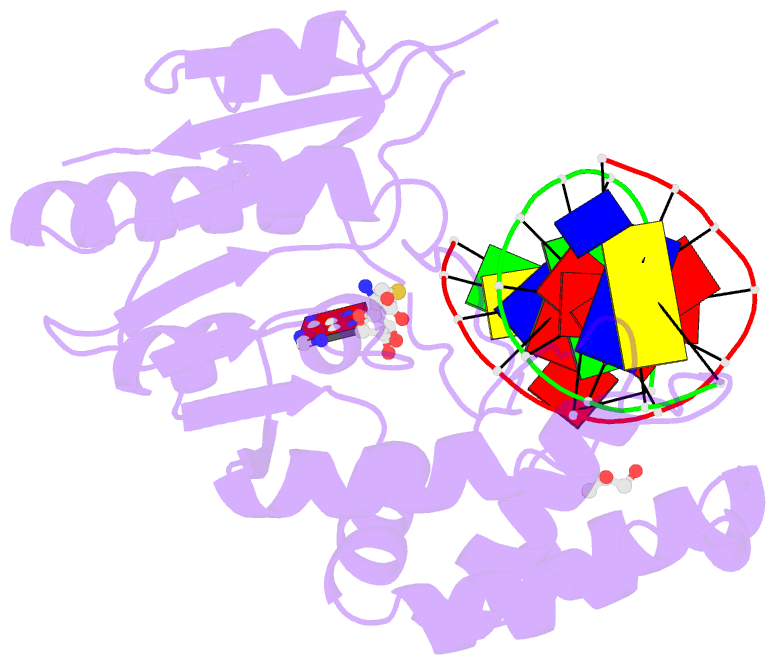
Summary information and primary citation
- PDB-id
- 4rtk; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (1.96 Å)
- Summary
- Complex of escherichia coli DNA adenine methyltransferase (dam) with sah and with DNA containing distal pap regulon sequence
- Reference
- Horton JR, Zhang X, Blumenthal RM, Cheng X (2015): "Structures of Escherichia coli DNA adenine methyltransferase (Dam) in complex with a non-GATC sequence: potential implications for methylation-independent transcriptional repression." Nucleic Acids Res., 43, 4296-4308. doi: 10.1093/nar/gkv251.
- Abstract
- DNA adenine methyltransferase (Dam) is widespread and conserved among the γ-proteobacteria. Methylation of the Ade in GATC sequences regulates diverse bacterial cell functions, including gene expression, mismatch repair and chromosome replication. Dam also controls virulence in many pathogenic Gram-negative bacteria. An unexplained and perplexing observation about Escherichia coli Dam (EcoDam) is that there is no obvious relationship between the genes that are transcriptionally responsive to Dam and the promoter-proximal presence of GATC sequences. Here, we demonstrate that EcoDam interacts with a 5-base pair non-cognate sequence distinct from GATC. The crystal structure of a non-cognate complex allowed us to identify a DNA binding element, GTYTA/TARAC (where Y = C/T and R = A/G). This element immediately flanks GATC sites in some Dam-regulated promoters, including the Pap operon which specifies pyelonephritis-associated pili. In addition, Dam interacts with near-cognate GATC sequences (i.e. 3/4-site ATC and GAT). Taken together, these results imply that Dam, in addition to being responsible for GATC methylation, could also function as a methylation-independent transcriptional repressor.