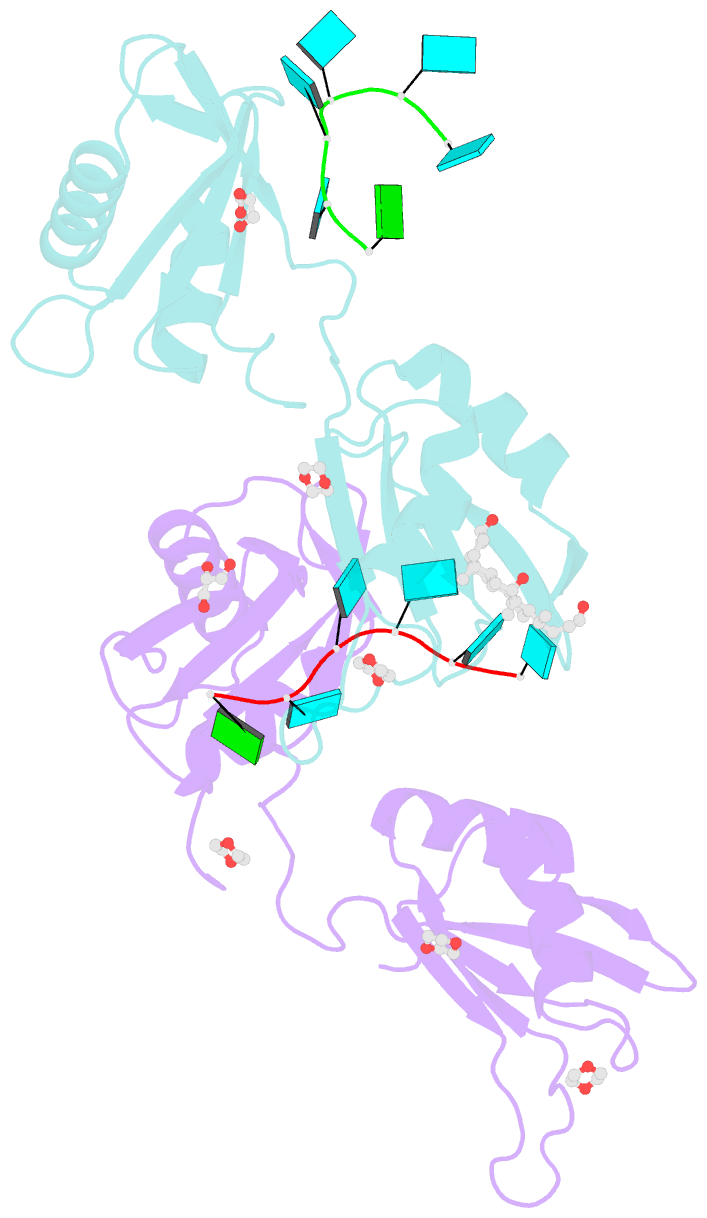
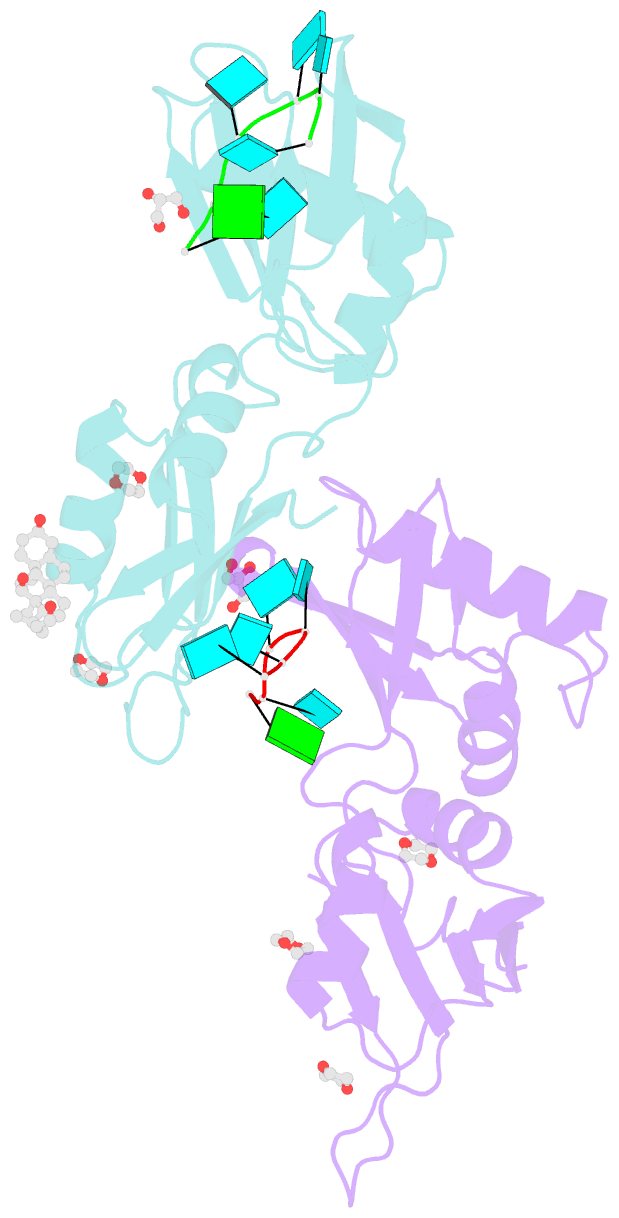
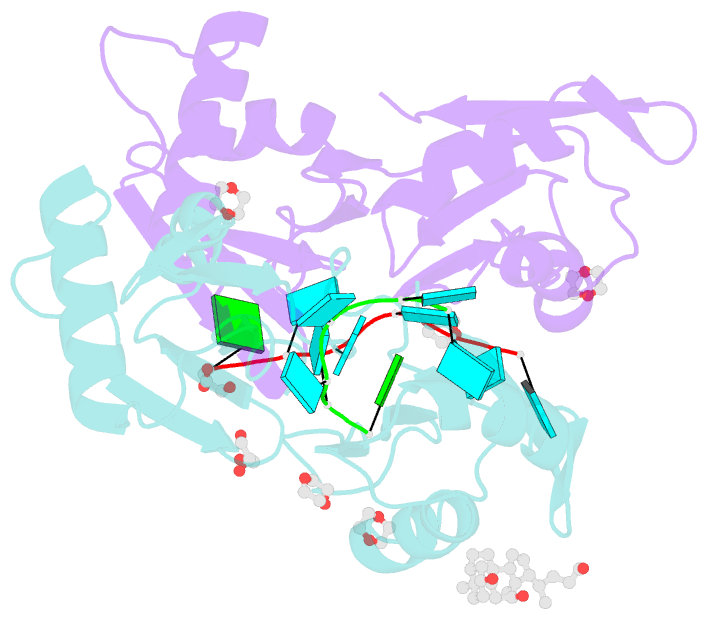
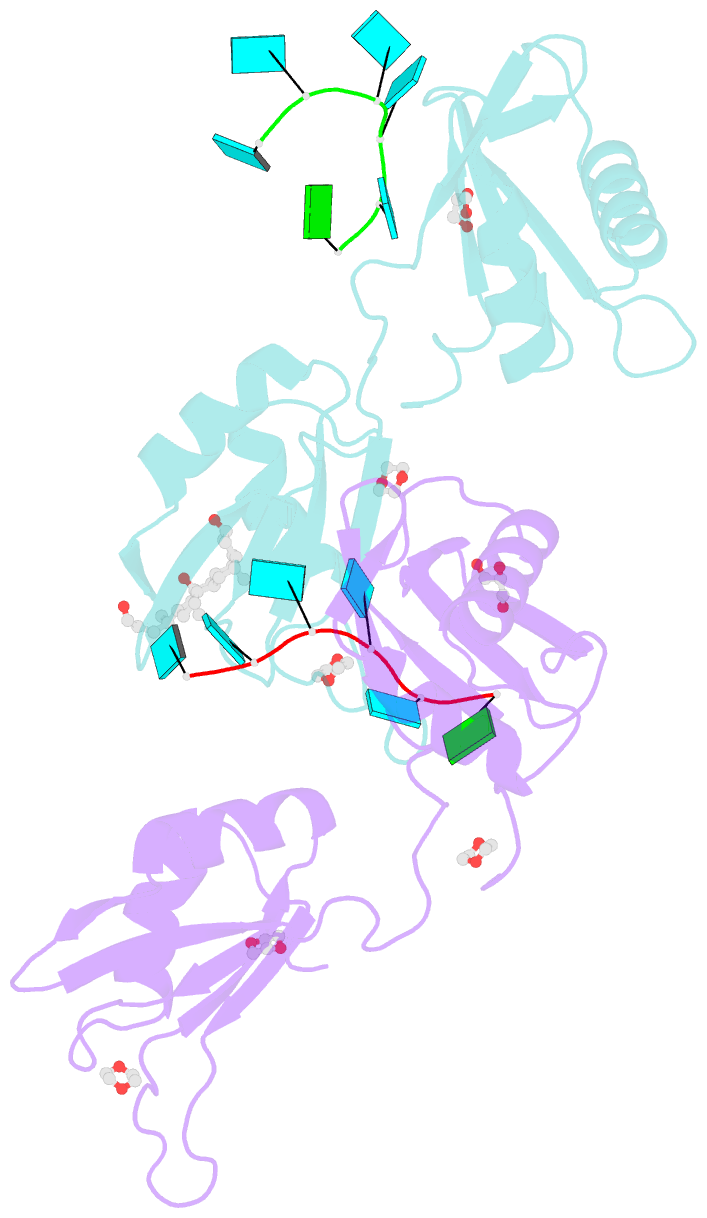
Summary information and primary citation
- PDB-id
- 4tu9; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-DNA
- Method
- X-ray (1.992 Å)
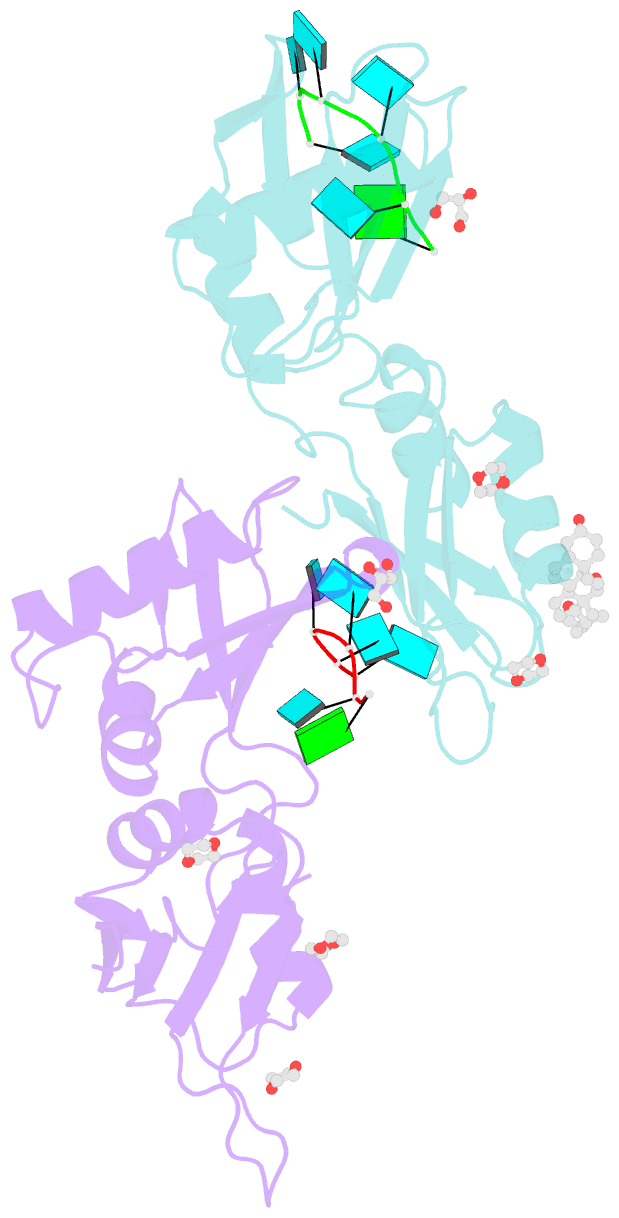
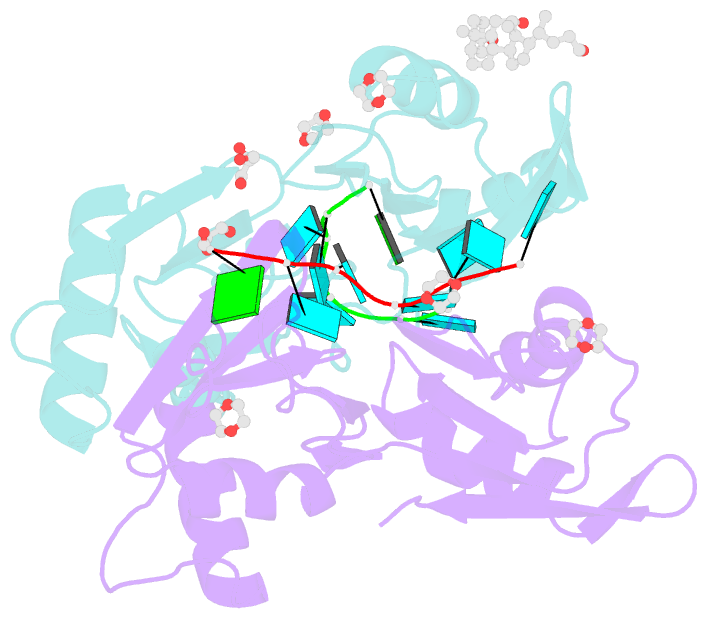
- Summary
- Structure of u2af65 variant with bru5g6 DNA
- Reference
- Agrawal AA, McLaughlin KJ, Jenkins JL, Kielkopf CL (2014): "Structure-guided U2AF65 variant improves recognition and splicing of a defective pre-mRNA." Proc.Natl.Acad.Sci.USA, 111, 17420-17425. doi: 10.1073/pnas.1412743111.
- Abstract
- Purine interruptions of polypyrimidine (Py) tract splice site signals contribute to human genetic diseases. The essential splicing factor U2AF(65) normally recognizes a Py tract consensus sequence preceding the major class of 3' splice sites. We found that neurofibromatosis- or retinitis pigmentosa-causing mutations in the 5' regions of Py tracts severely reduce U2AF(65) affinity. Conversely, we identified a preferred binding site of U2AF(65) for purine substitutions in the 3' regions of Py tracts. Based on a comparison of new U2AF(65) structures bound to either A- or G-containing Py tracts with previously identified pyrimidine-containing structures, we expected to find that a D231V amino acid change in U2AF(65) would specify U over other nucleotides. We found that the crystal structure of the U2AF(65)-D231V variant confirms favorable packing between the engineered valine and a target uracil base. The D231V amino acid change restores U2AF(65) affinity for two mutated splice sites that cause human genetic diseases and successfully promotes splicing of a defective retinitis pigmentosa-causing transcript. We conclude that reduced U2AF(65) binding is a molecular consequence of disease-relevant mutations, and that a structure-guided U2AF(65) variant is capable of manipulating gene expression in eukaryotic cells.