Summary information and primary citation
- PDB-id
- 4tug; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (3.55 Å)
- Summary
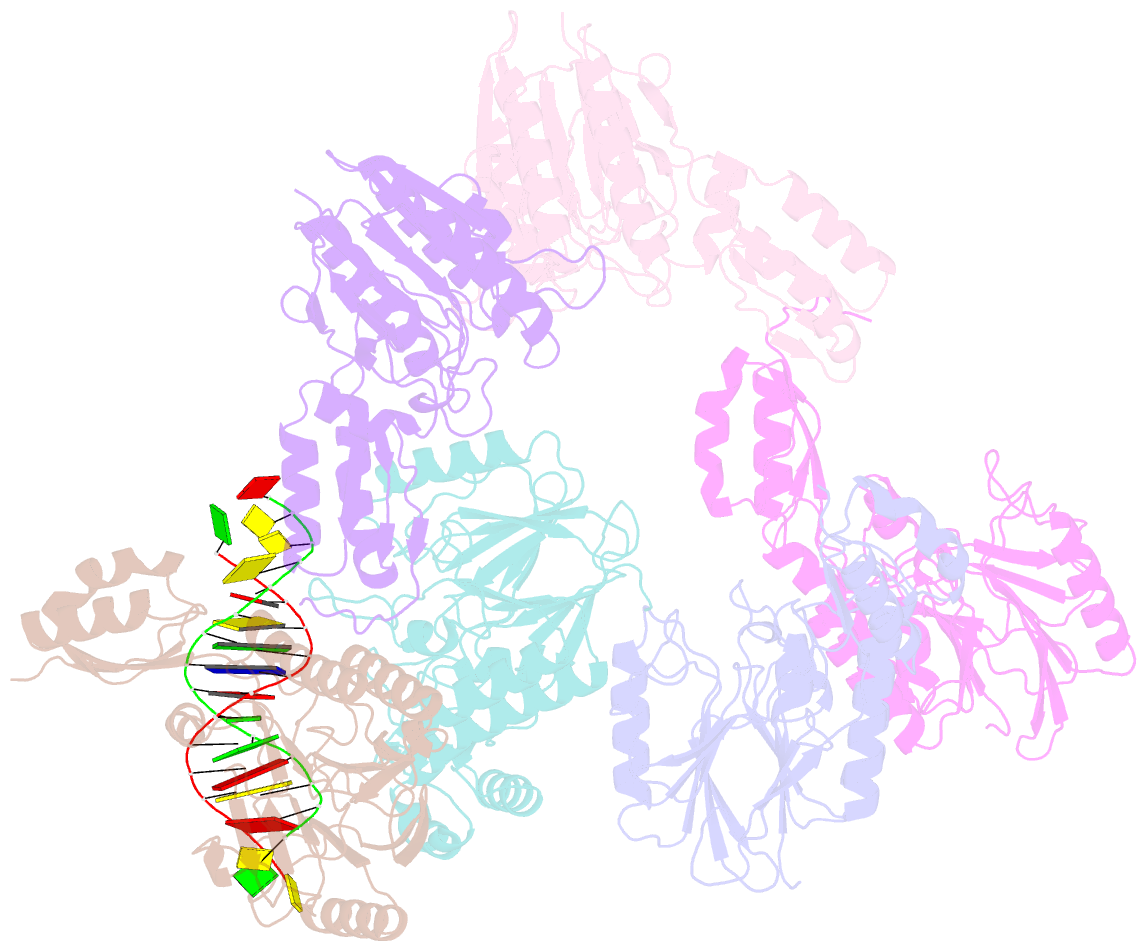
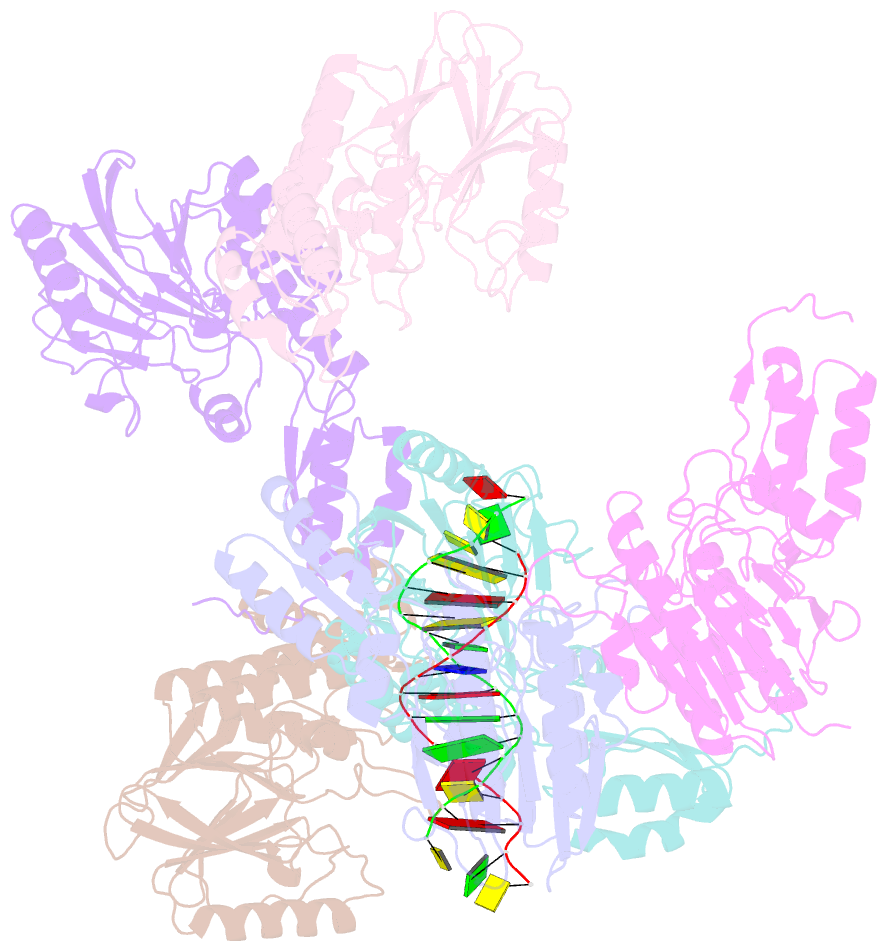
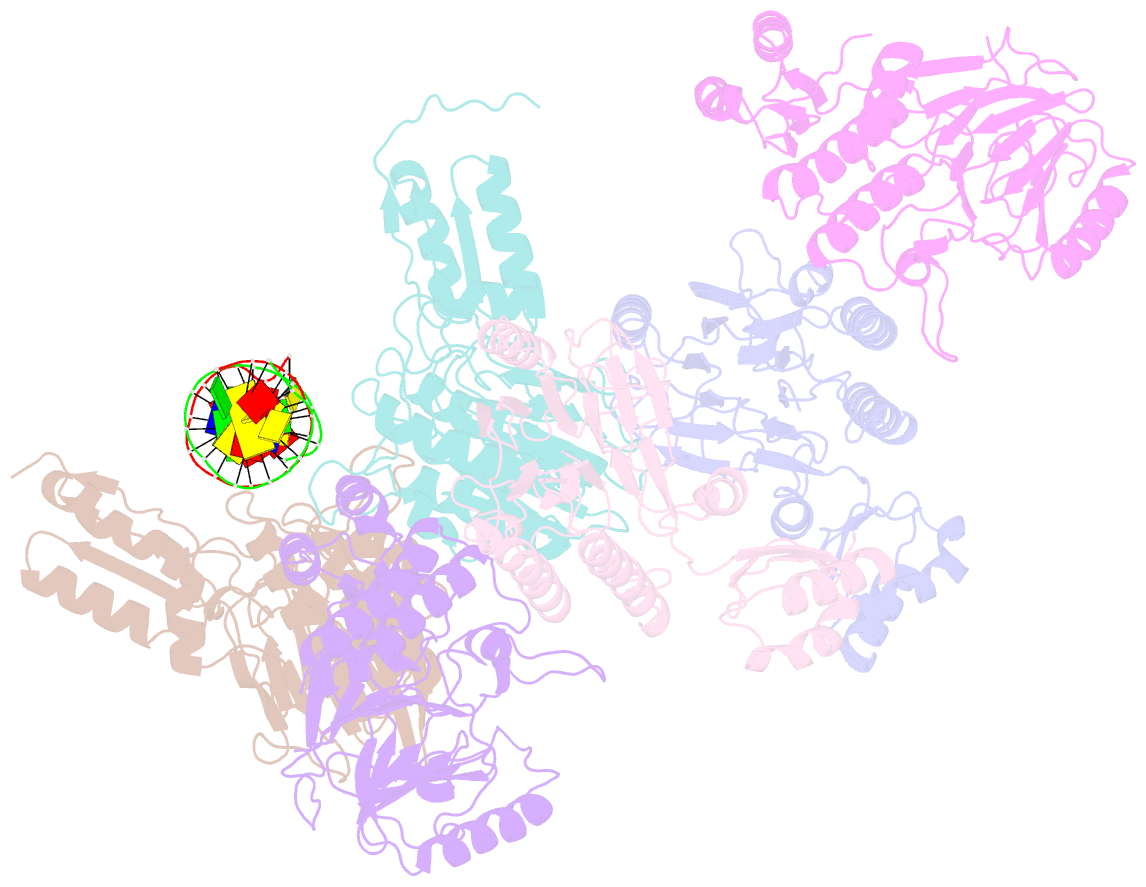
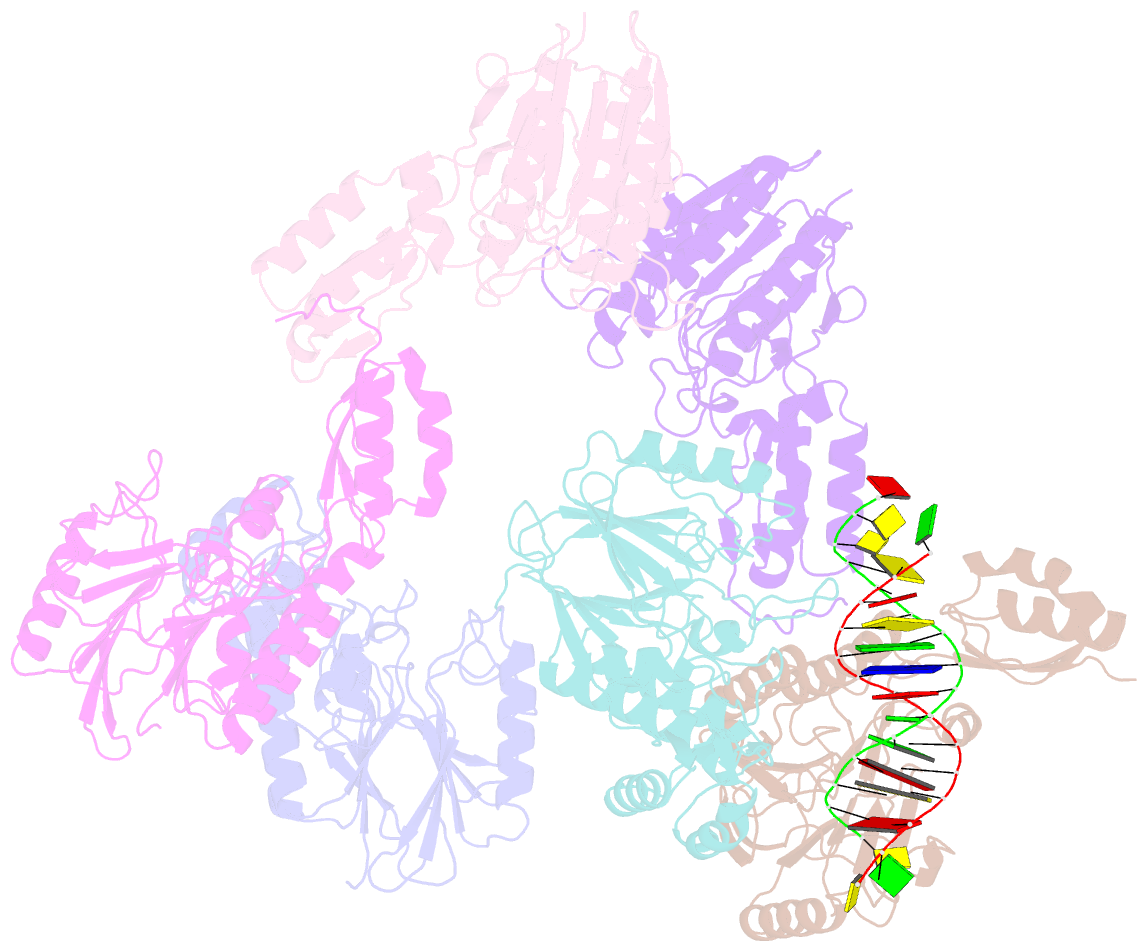
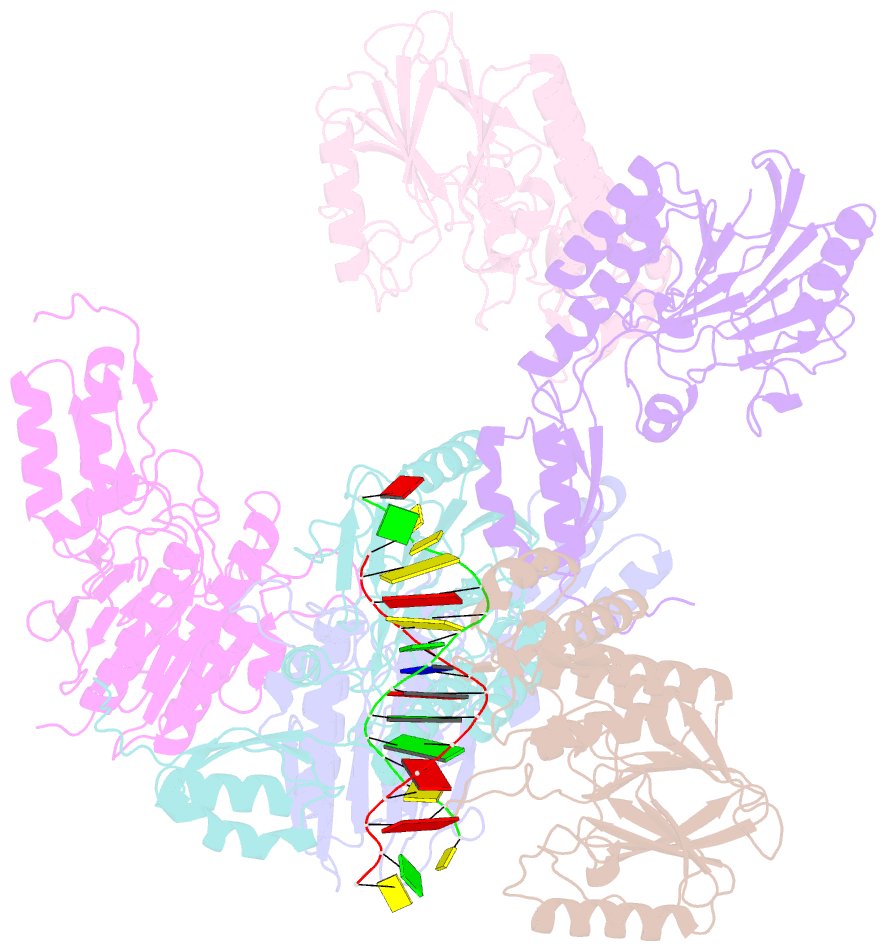
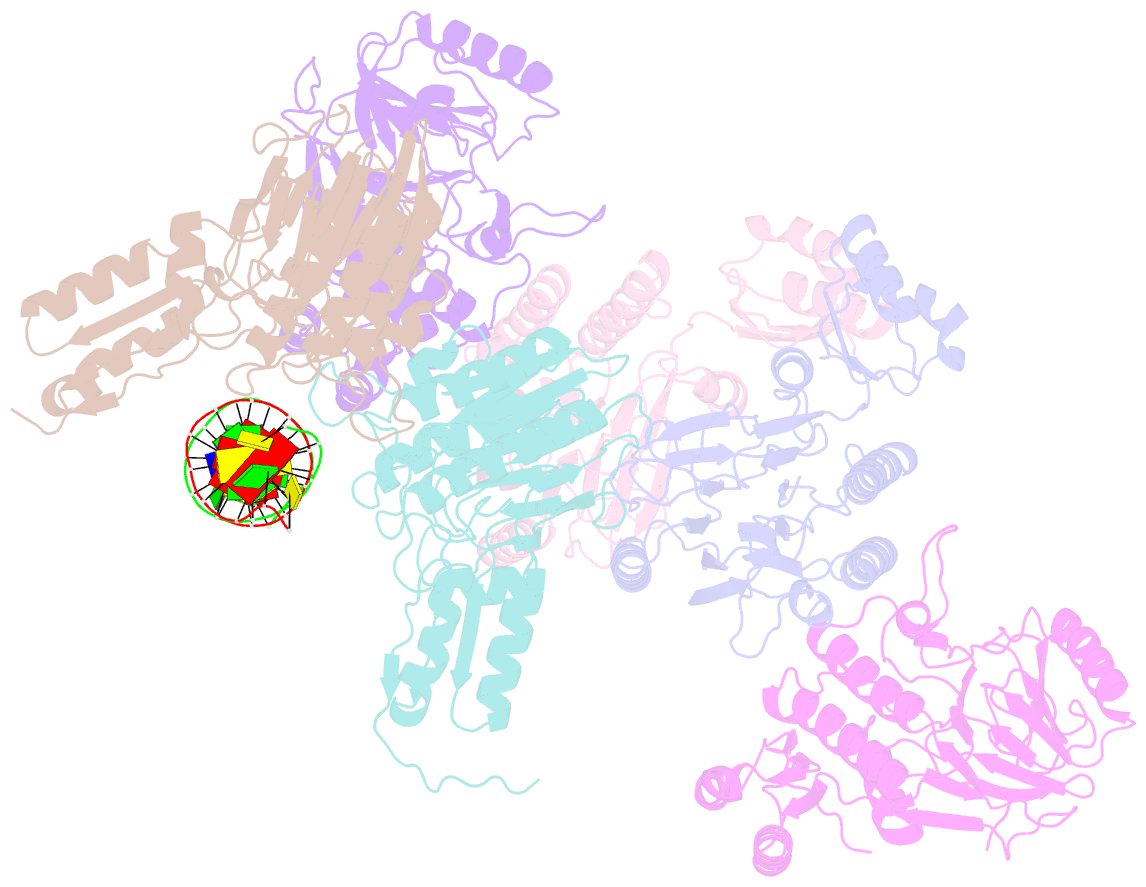
- Crystal structure of mjmre11-dna2 complex
- Reference
- Sung S, Li F, Park YB, Kim JS, Kim AK, Song OK, Kim J, Che J, Lee SE, Cho Y (2014): "DNA end recognition by the Mre11 nuclease dimer: insights into resection and repair of damaged DNA." Embo J., 33, 2422-2435. doi: 10.15252/embj.201488299.
- Abstract
- The Mre11-Rad50-Nbs1 (MRN) complex plays important roles in sensing DNA damage, as well as in resecting and tethering DNA ends, and thus participates in double-strand break repair. An earlier structure of Mre11 bound to a short duplex DNA molecule suggested that each Mre11 in a dimer recognizes one DNA duplex to bridge two DNA ends at a short distance. Here, we provide an alternative DNA recognition model based on the structures of Methanococcus jannaschii Mre11 (MjMre11) bound to longer DNA molecules, which may more accurately reflect a broken chromosome. An extended stretch of B-form DNA asymmetrically runs across the whole dimer, with each end of this DNA molecule being recognized by an individual Mre11 monomer. DNA binding induces rigid-body rotation of the Mre11 dimer, which could facilitate melting of the DNA end and its juxtaposition to an active site of Mre11. The identified Mre11 interface binding DNA duplex ends is structurally conserved and shown to functionally contribute to efficient resection, non-homologous end joining, and tolerance to DNA-damaging agents when other resection enzymes are absent. Together, the structural, biochemical, and genetic findings presented here offer new insights into how Mre11 recognizes damaged DNA and facilitates DNA repair.