Summary information and primary citation
- PDB-id
- 4u0y; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (1.91 Å)
- Summary
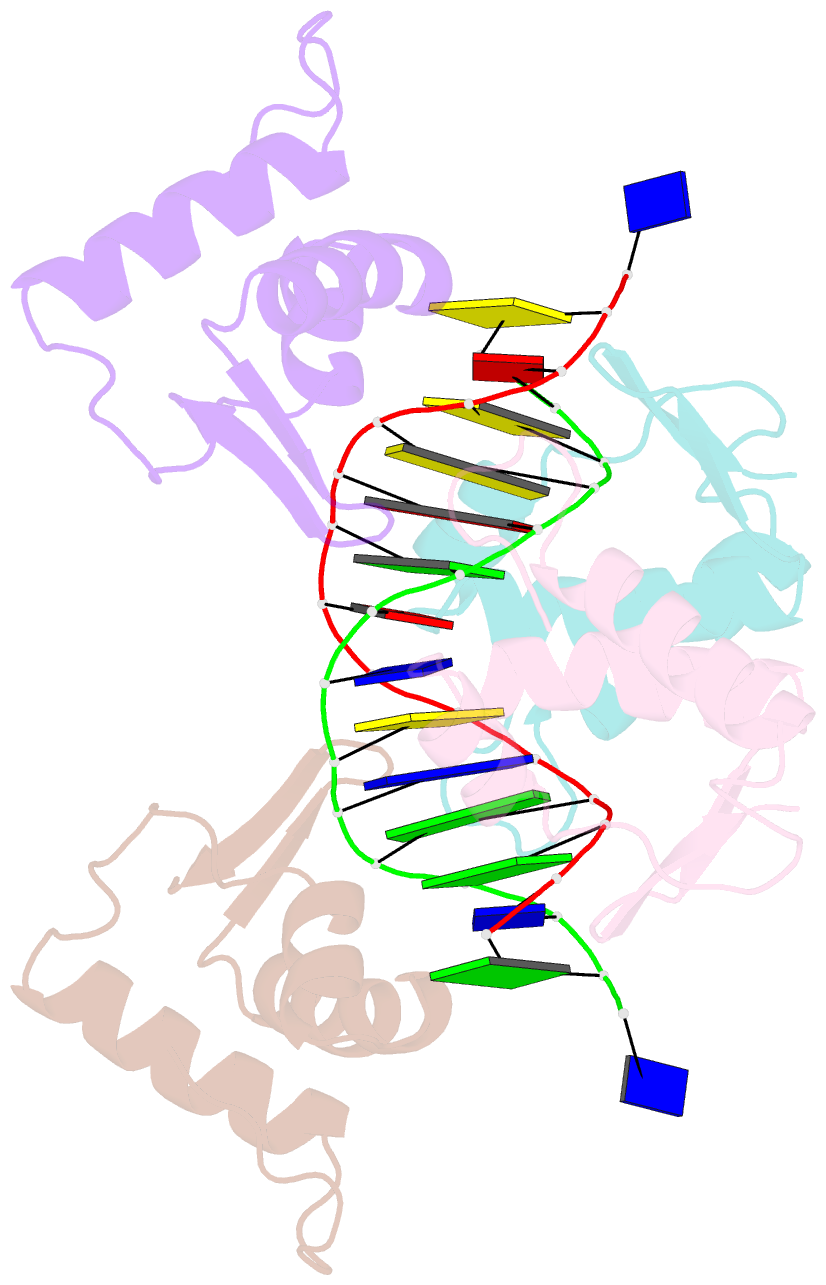
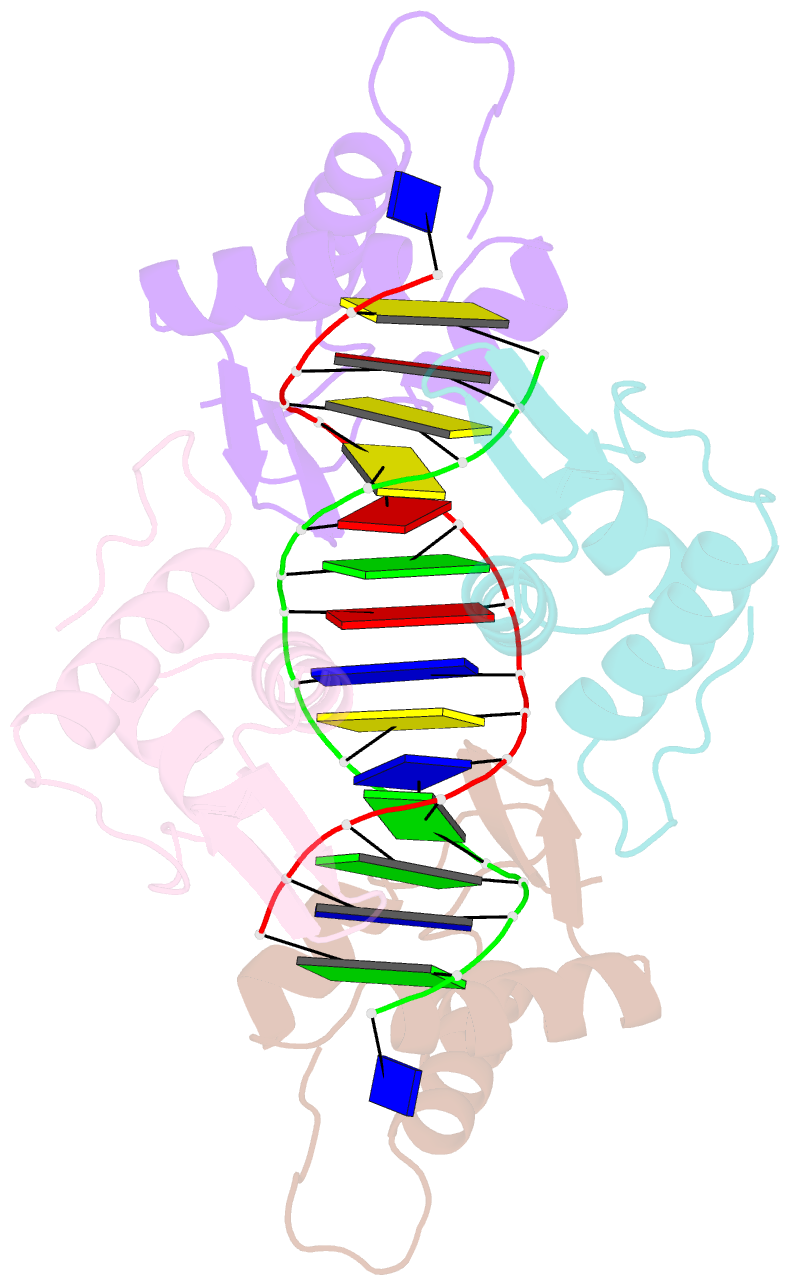
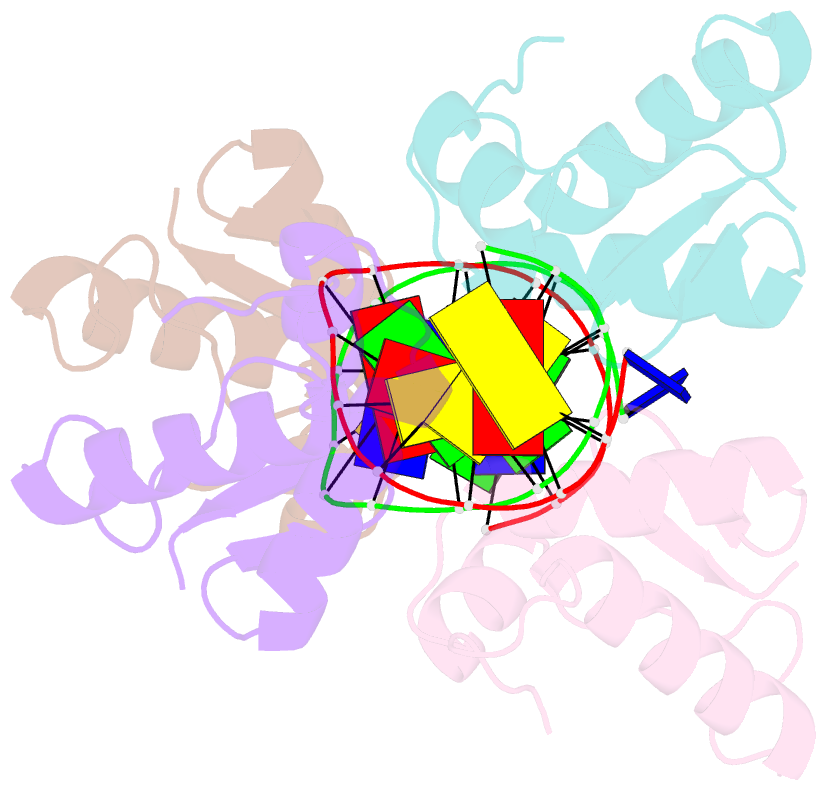
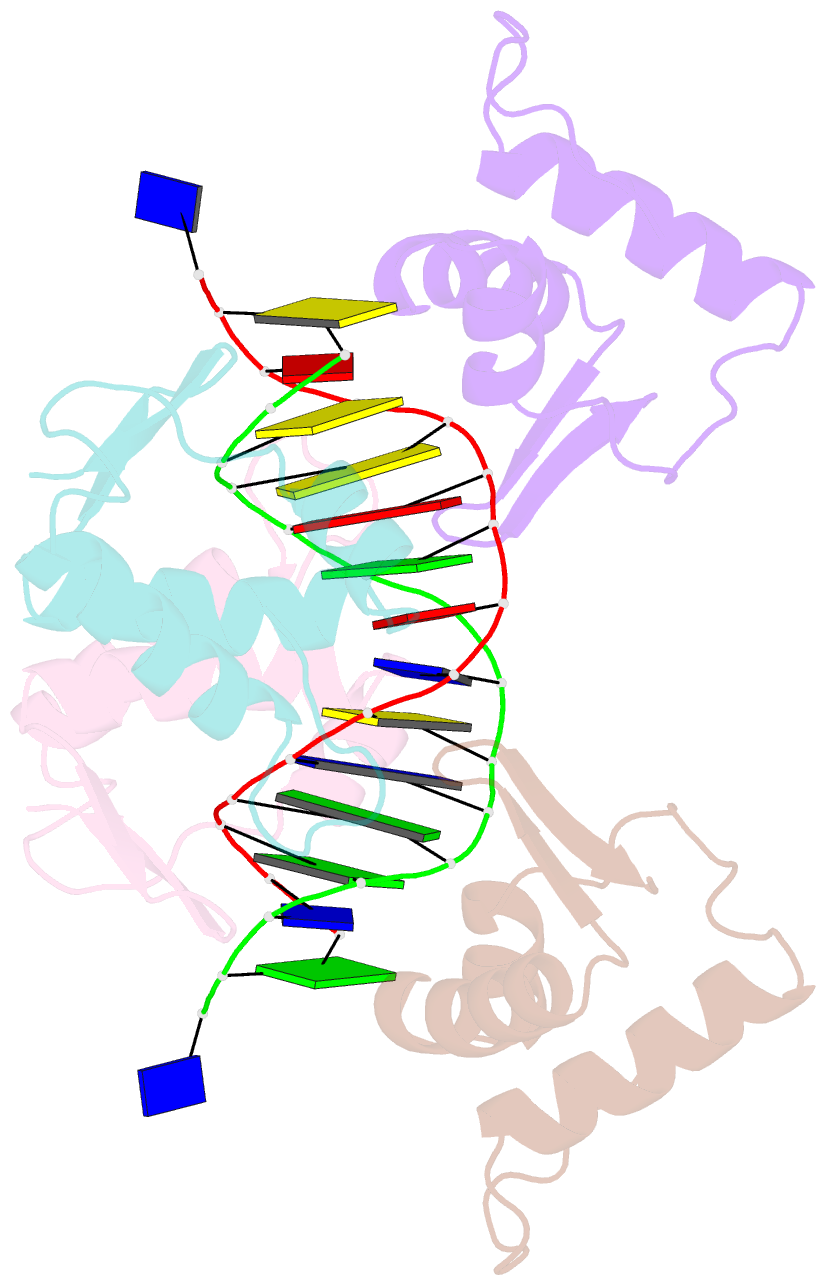
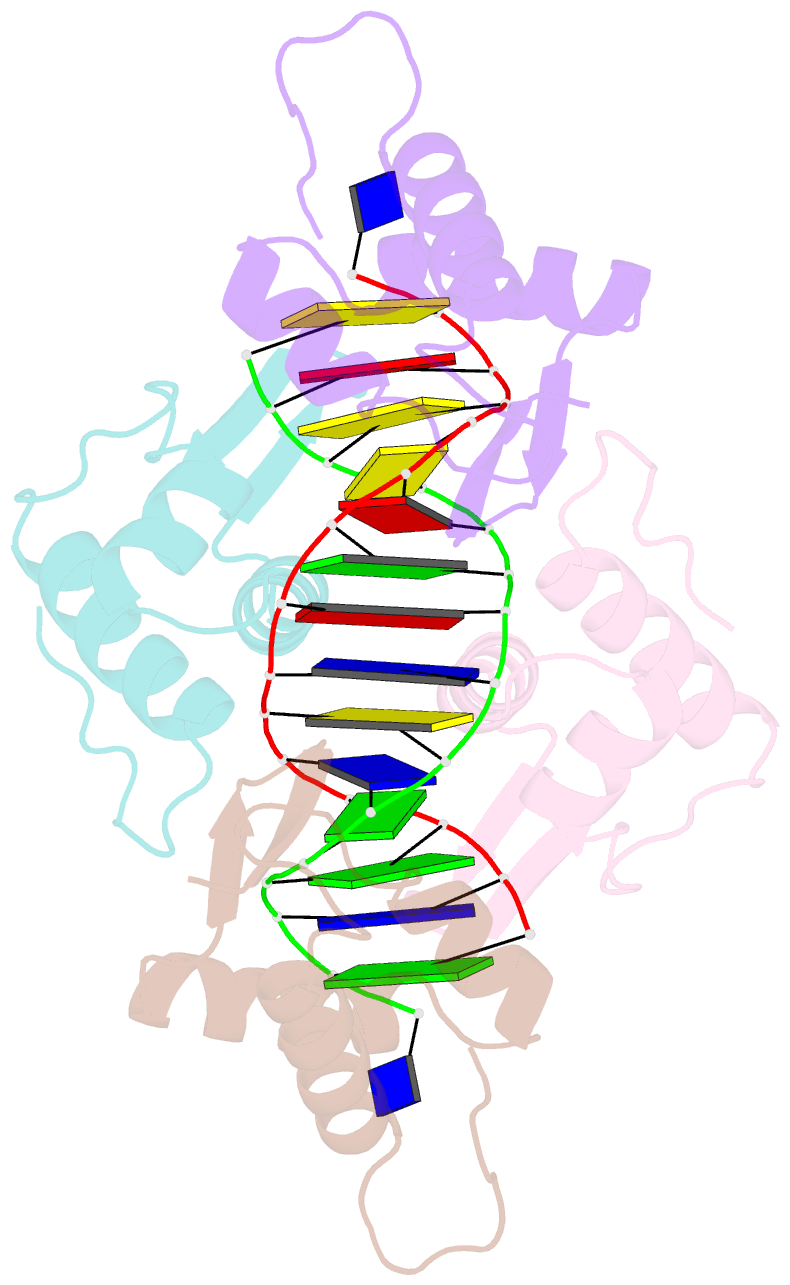
- Crystal structure of the DNA-binding domains of yvoa in complex with palindromic operator DNA
- Reference
- Fillenberg SB, Grau FC, Seidel G, Muller YA (2015): "Structural insight into operator dre-sites recognition and effector binding in the GntR/HutC transcription regulator NagR." Nucleic Acids Res., 43, 1283-1296. doi: 10.1093/nar/gku1374.
- Abstract
- The uptake and metabolism of N-acetylglucosamine (GlcNAc) in Bacillus subtilis is controlled by NagR (formerly named YvoA), a member of the widely-occurring GntR/HutC family of transcription regulators. Upon binding to specific DNA operator sites (dre-sites) NagR blocks the transcription of genes for GlcNAc utilization and interaction of NagR with effectors abrogates gene repression. Here we report crystal structures of NagR in complex with operator DNA and in complex with the putative effector molecules glucosamine-6-phosphate (GlcN-6-P) and N-acetylglucosamine-6-phosphate (GlcNAc-6-P). A comparison of the distinct conformational states suggests that effectors are able to displace the NagR-DNA-binding domains (NagR-DBDs) by almost 70 Å upon binding. In addition, a high-resolution crystal structure of isolated NagR-DBDs in complex with palindromic double-stranded DNA (dsDNA) discloses both the determinants for highly sequence-specific operator dre-site recognition and for the unspecific binding of NagR to dsDNA. Extensive biochemical binding studies investigating the affinities of full-length NagR and isolated NagR-DBDs for either random DNA, dre-site-derived palindromic or naturally occurring non-palindromic dre-site sequences suggest that proper NagR function relies on an effector-induced fine-tuning of the DNA-binding affinities of NagR and not on a complete abrogation of its DNA binding.