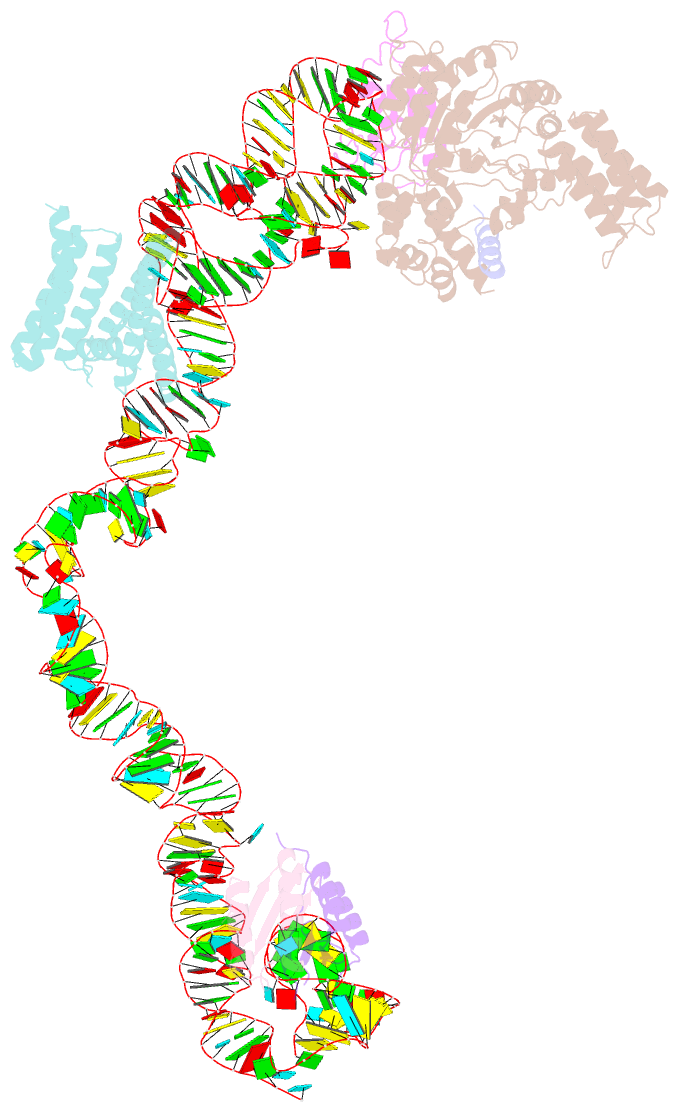
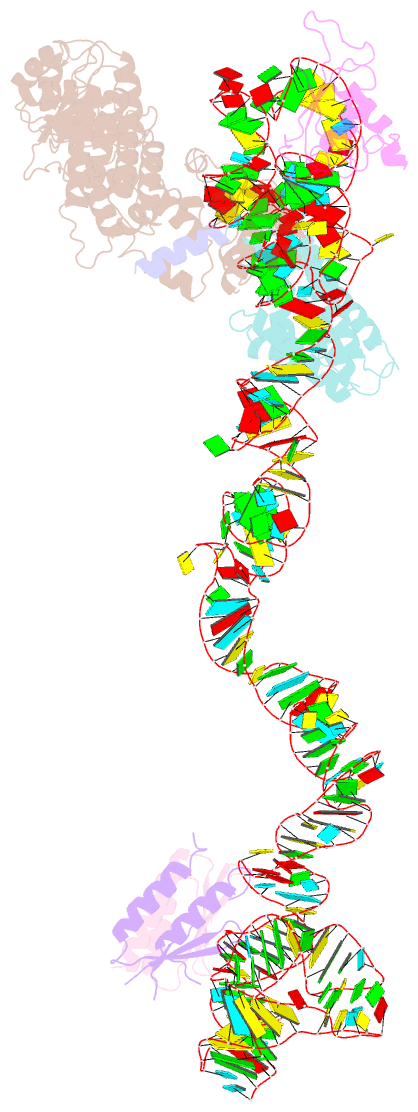
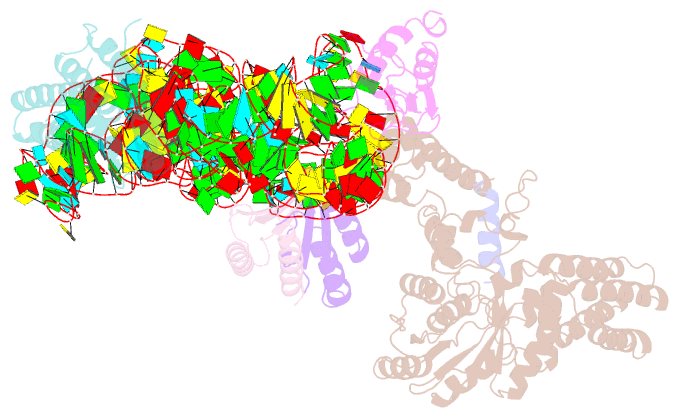
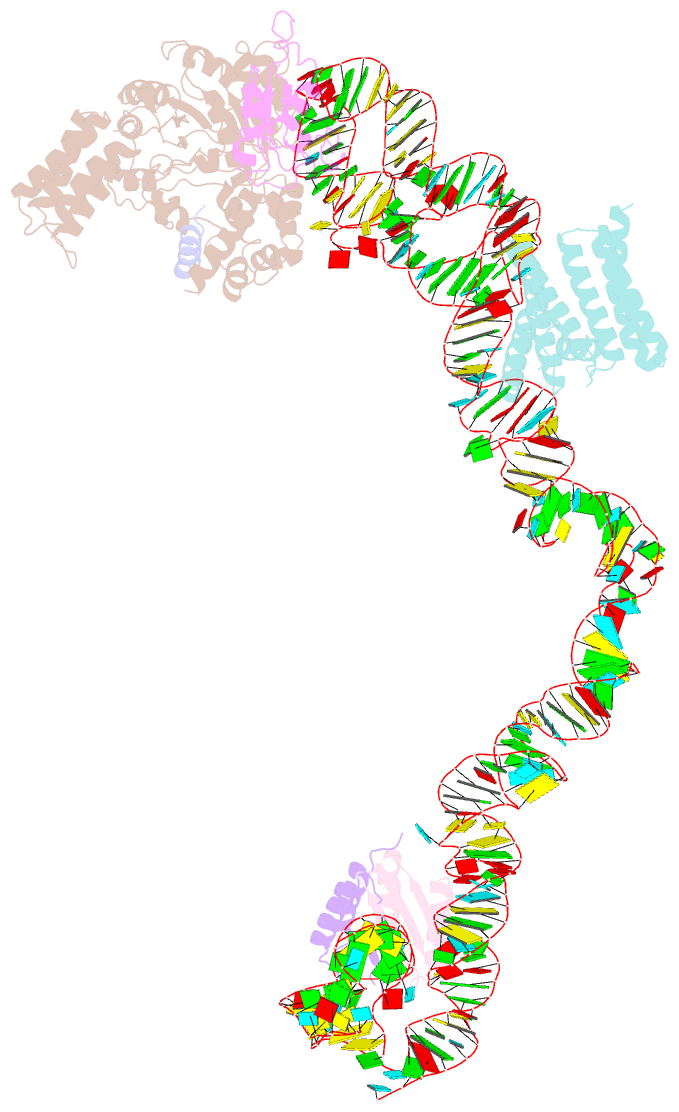
Summary information and primary citation
- PDB-id
- 4ue5; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- translation
- Method
- cryo-EM (9.0 Å)
- Summary
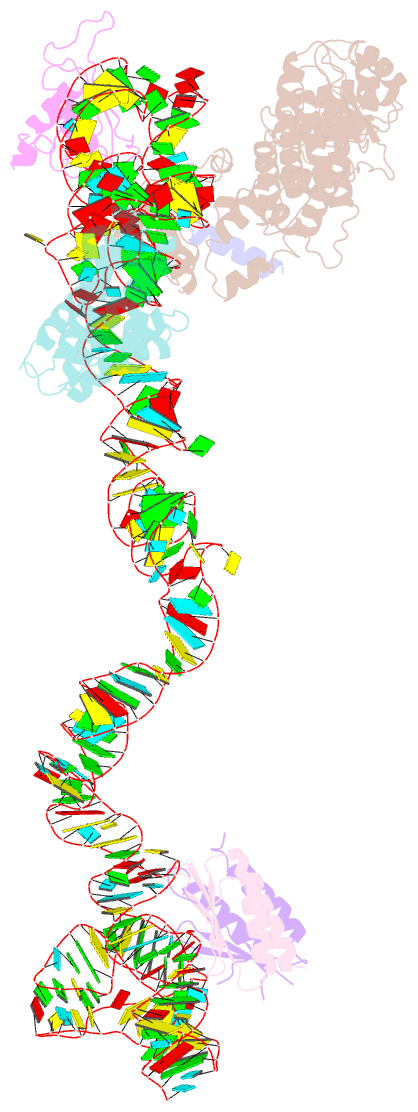
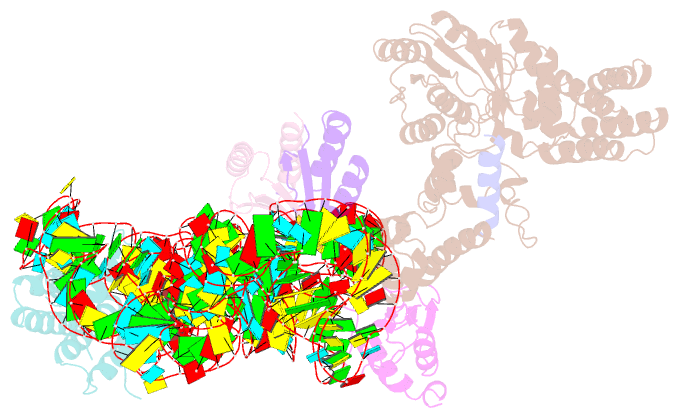
- Structural basis for targeting and elongation arrest of bacillus signal recognition particle
- Reference
- Beckert B, Kedrov A, Sohmen D, Kempf G, Wild K, Sinning I, Stahlberg H, Wilson DN, Beckmann R (2015): "Translational Arrest by a Prokaryotic Signal Recognition Particle is Mediated by RNA Interactions." Nat.Struct.Mol.Biol., 22, 767. doi: 10.1038/NSMB.3086.
- Abstract
- The signal recognition particle (SRP) recognizes signal sequences of nascent polypeptides and targets ribosome-nascent chain complexes to membrane translocation sites. In eukaryotes, translating ribosomes are slowed down by the Alu domain of SRP to allow efficient targeting. In prokaryotes, however, little is known about the structure and function of Alu domain-containing SRPs. Here, we report a complete molecular model of SRP from the Gram-positive bacterium Bacillus subtilis, based on cryo-EM. The SRP comprises two subunits, 6S RNA and SRP54 or Ffh, and it facilitates elongation slowdown similarly to its eukaryotic counterpart. However, protein contacts with the small ribosomal subunit observed for the mammalian Alu domain are substituted in bacteria by RNA-RNA interactions of 6S RNA with the α-sarcin-ricin loop and helices H43 and H44 of 23S rRNA. Our findings provide a structural basis for cotranslational targeting and RNA-driven elongation arrest in prokaryotes.