Summary information and primary citation
- PDB-id
- 4uyk; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- signaling protein
- Method
- X-ray (3.22 Å)
- Summary
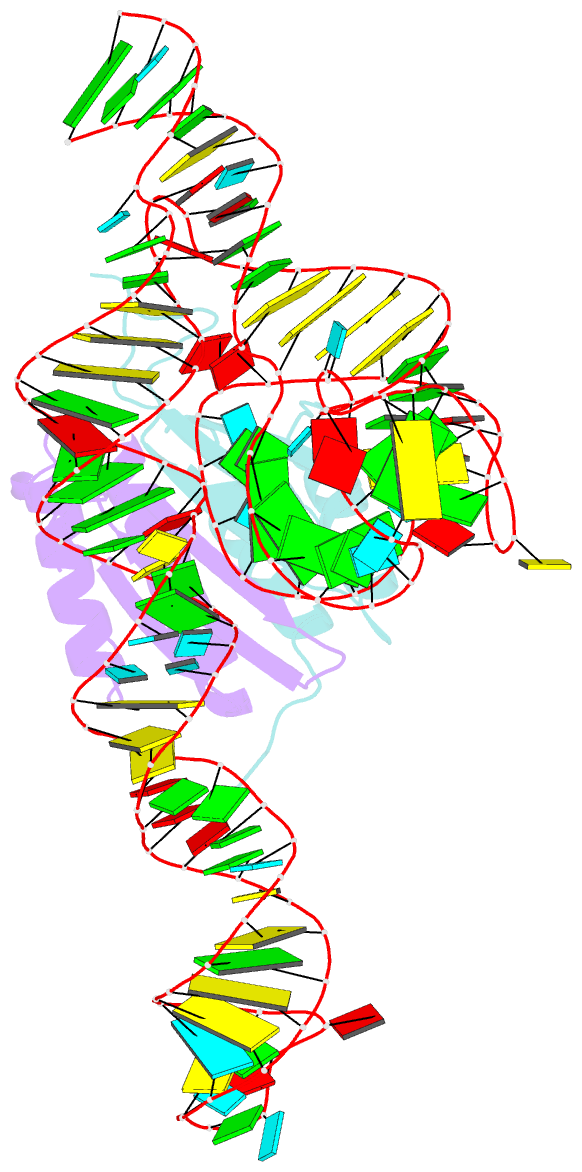
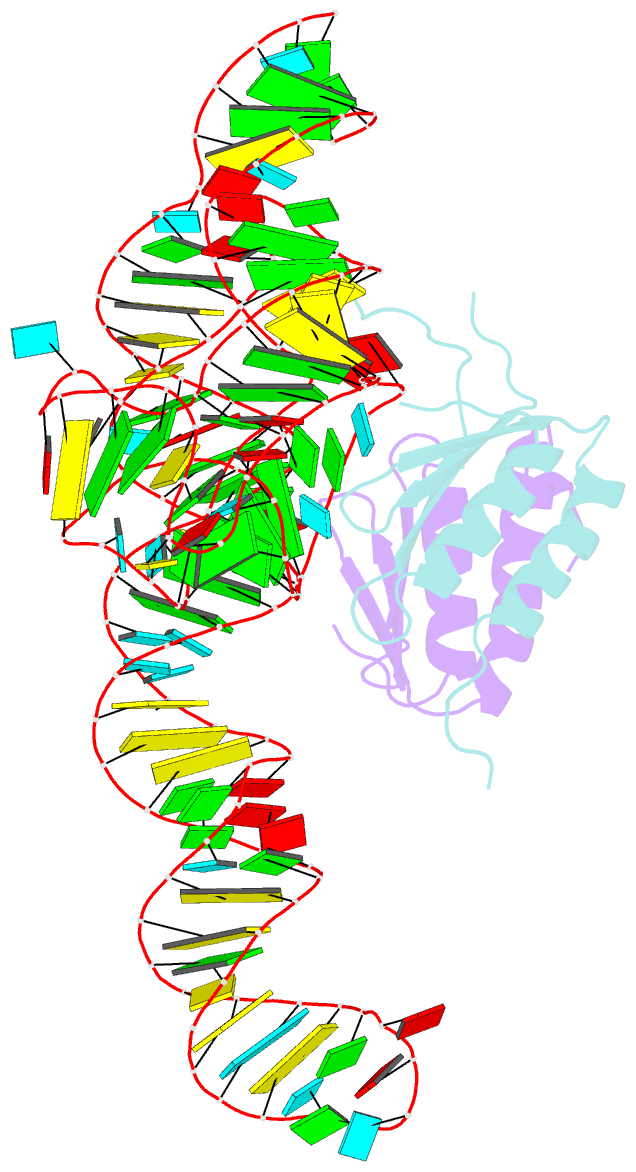
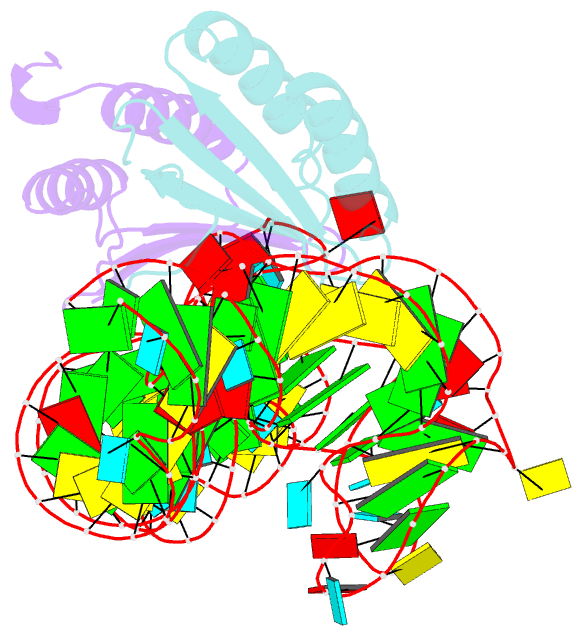
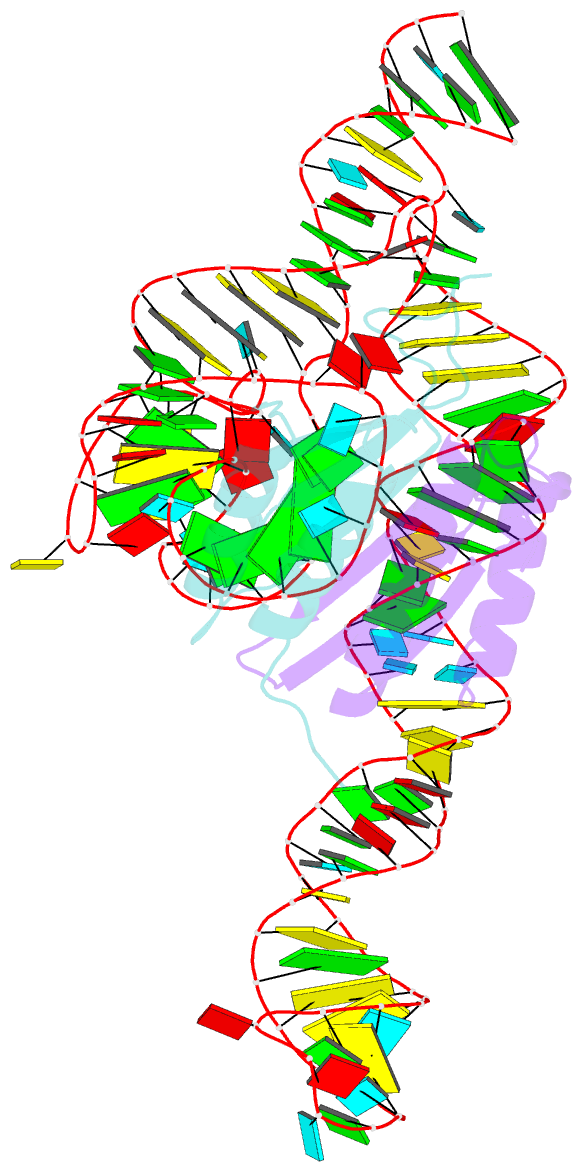
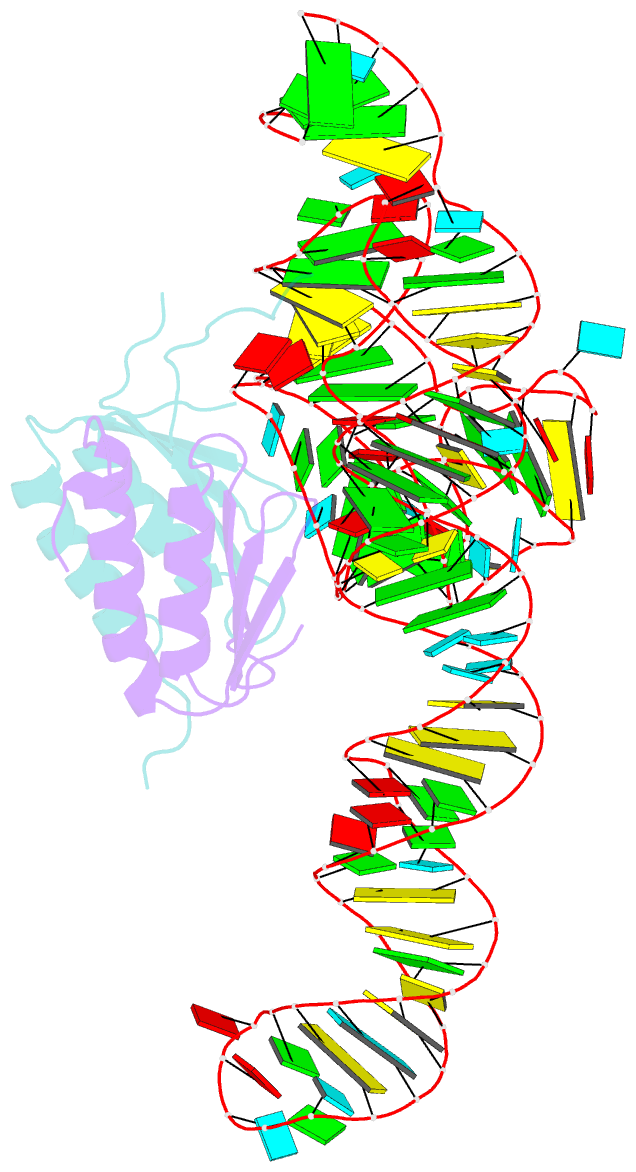
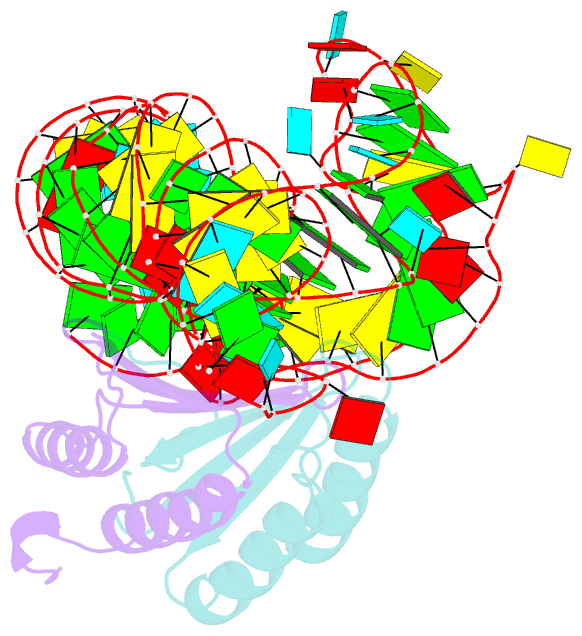
- Crystal structure of a signal recognition particle alu domain in the elongation arrest conformation
- Reference
- Bousset L, Mary C, Brooks MA, Scherrer A, Strub K, Cusack S (2014): "Crystal Structure of a Signal Recognition Particle Alu Domain in the Elongation Arrest Conformation." RNA, 20, 1955. doi: 10.1261/RNA.047209.114.
- Abstract
- The signal recognition particle (SRP) is a conserved ribonucleoprotein particle that targets membrane and secreted proteins to translocation channels in membranes. In eukaryotes, the Alu domain, which comprises the 5' and 3' extremities of the SRP RNA bound to the SRP9/14 heterodimer, is thought to interact with the ribosome to pause translation elongation during membrane docking. We present the 3.2 Å resolution crystal structure of a chimeric Alu domain, comprising Alu RNA from the archaeon Pyrococcus horikoshii bound to the human Alu binding proteins SRP9/14. The structure reveals how intricate tertiary interactions stabilize the RNA 5' domain structure and how an extra, archaeal-specific, terminal stem helps constrain the Alu RNA into the active closed conformation. In this conformation, highly conserved noncanonical base pairs allow unusually tight side-by-side packing of 5' and 3' RNA stems within the SRP9/14 RNA binding surface. The biological relevance of this structure is confirmed by showing that a reconstituted full-length chimeric archaeal-human SRP is competent to elicit elongation arrest in vitro. The structure will be useful in refining our understanding of how the SRP Alu domain interacts with the ribosome.