Summary information and primary citation
- PDB-id
- 4w9m; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (2.7 Å)
- Summary
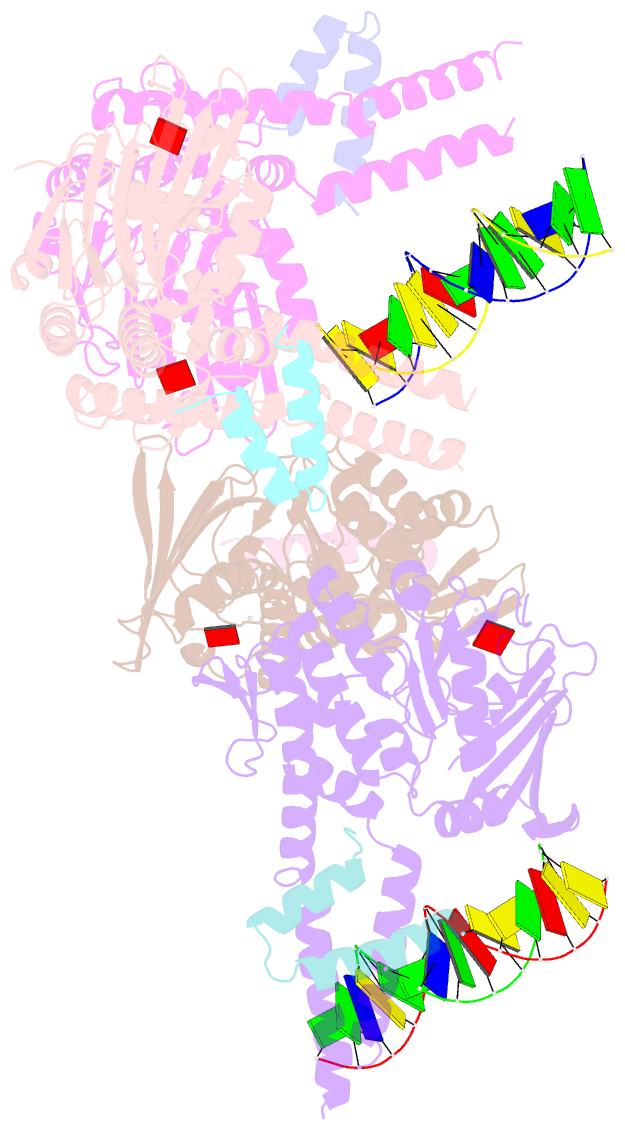
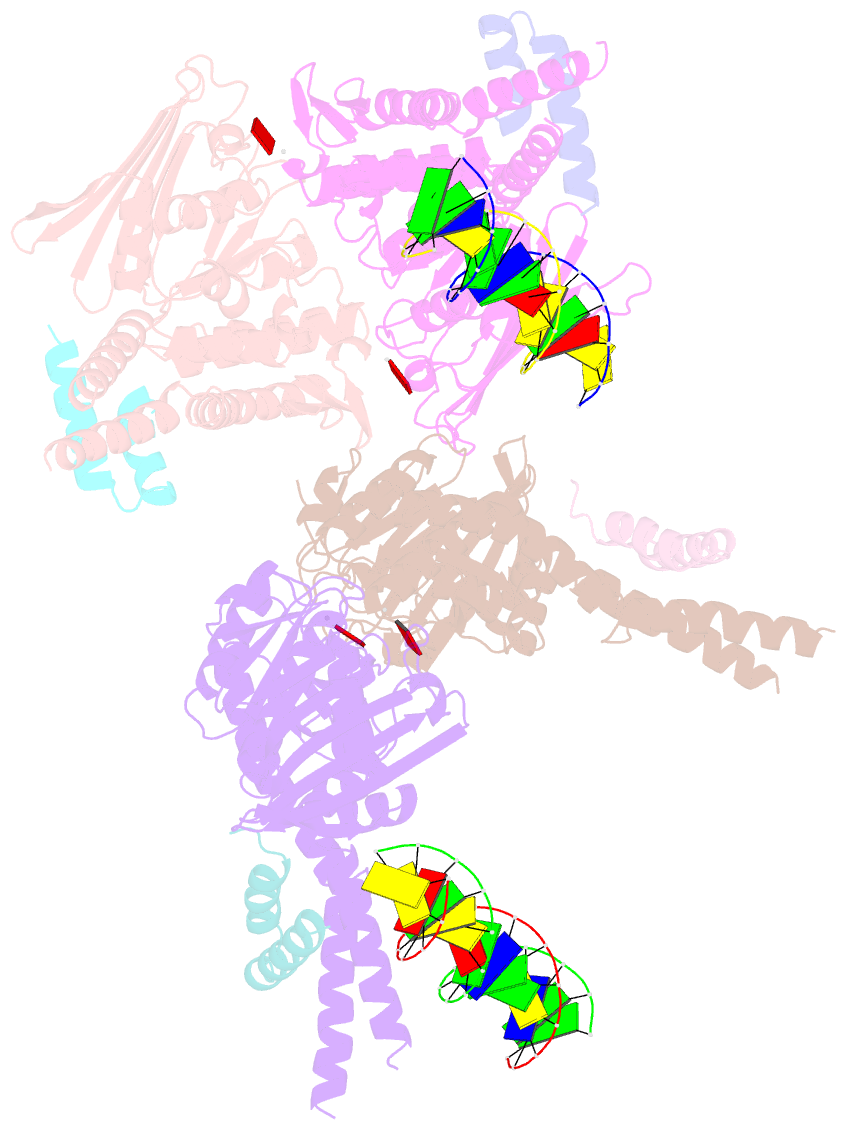
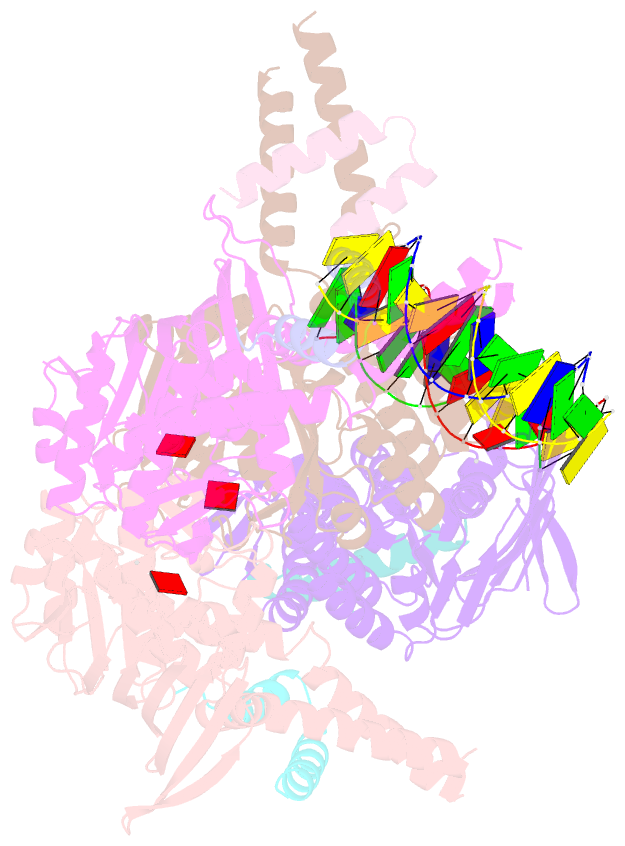
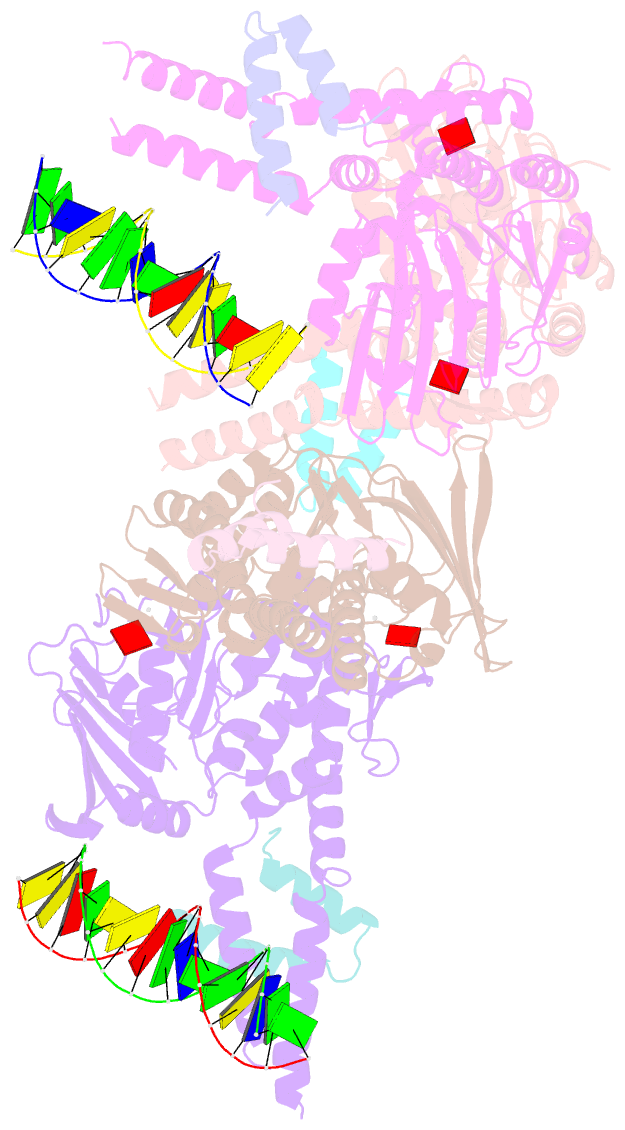
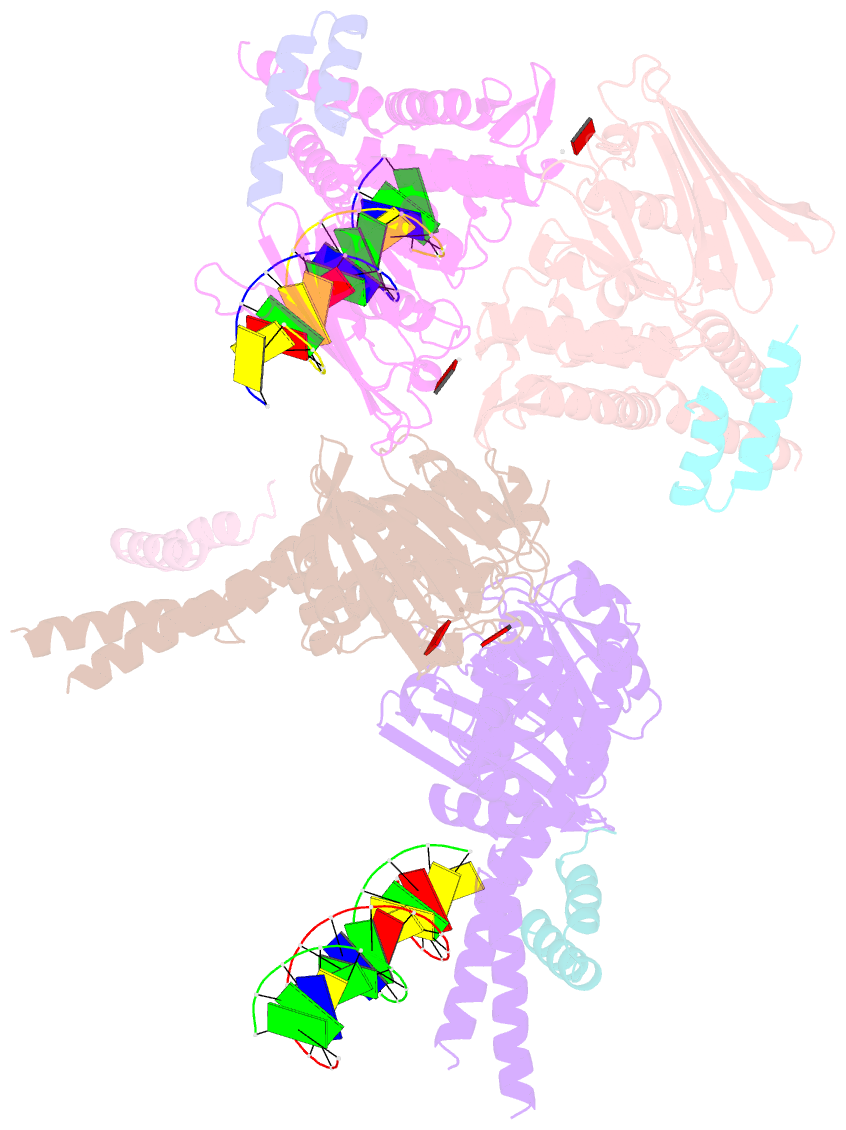
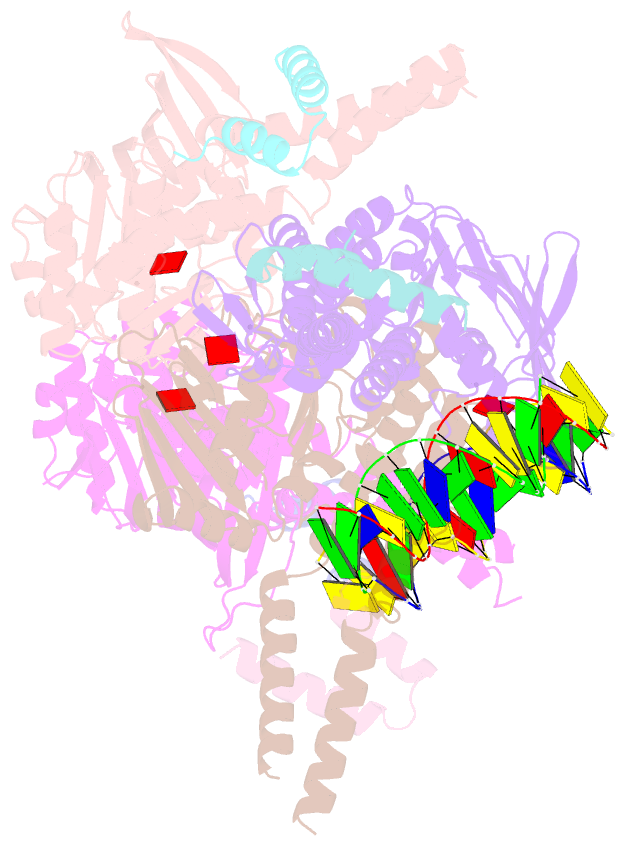
- Amppnp bound rad50 in complex with dsDNA
- Reference
- Rojowska A, Lammens K, Seifert FU, Direnberger C, Feldmann H, Hopfner KP (2014): "Structure of the Rad50 DNA double-strand break repair protein in complex with DNA." Embo J., 33, 2847-2859. doi: 10.15252/embj.201488889.
- Abstract
- The Mre11-Rad50 nuclease-ATPase is an evolutionarily conserved multifunctional DNA double-strand break (DSB) repair factor. Mre11-Rad50's mechanism in the processing, tethering, and signaling of DSBs is unclear, in part because we lack a structural framework for its interaction with DNA in different functional states. We determined the crystal structure of Thermotoga maritima Rad50(NBD) (nucleotide-binding domain) in complex with Mre11(HLH) (helix-loop-helix domain), AMPPNP, and double-stranded DNA. DNA binds between both coiled-coil domains of the Rad50 dimer with main interactions to a strand-loop-helix motif on the NBD. Our analysis suggests that this motif on Rad50 does not directly recognize DNA ends and binds internal sites on DNA. Functional studies reveal that DNA binding to Rad50 is not critical for DNA double-strand break repair but is important for telomere maintenance. In summary, we provide a structural framework for DNA binding to Rad50 in the ATP-bound state.