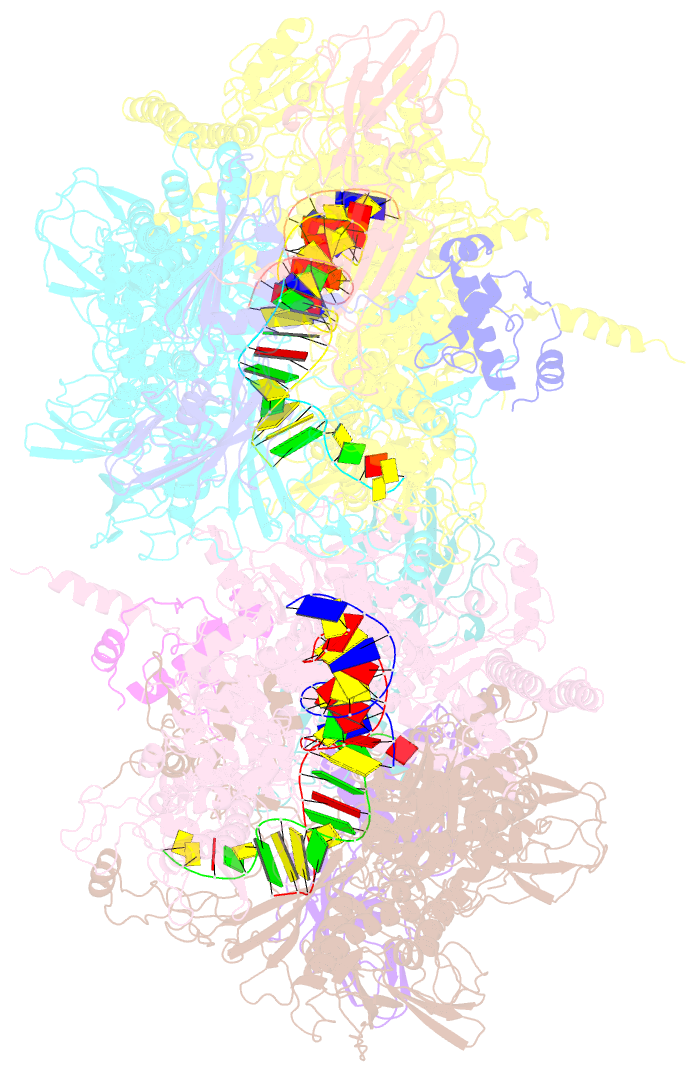
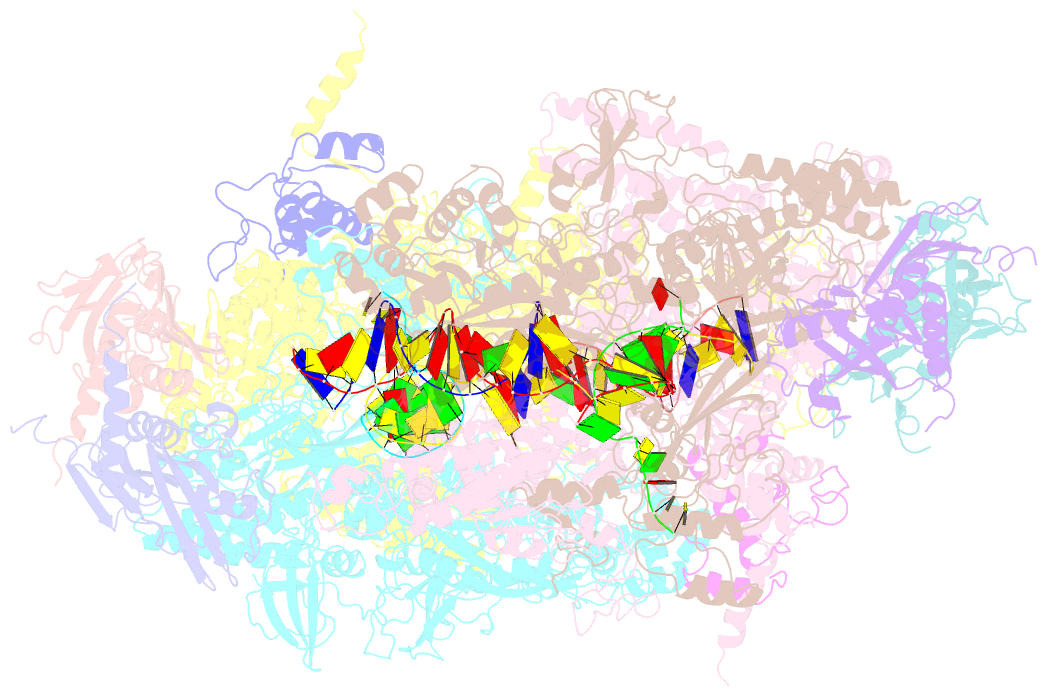
Summary information and primary citation
- PDB-id
- 4wqs; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA-RNA
- Method
- X-ray (4.306 Å)
- Summary
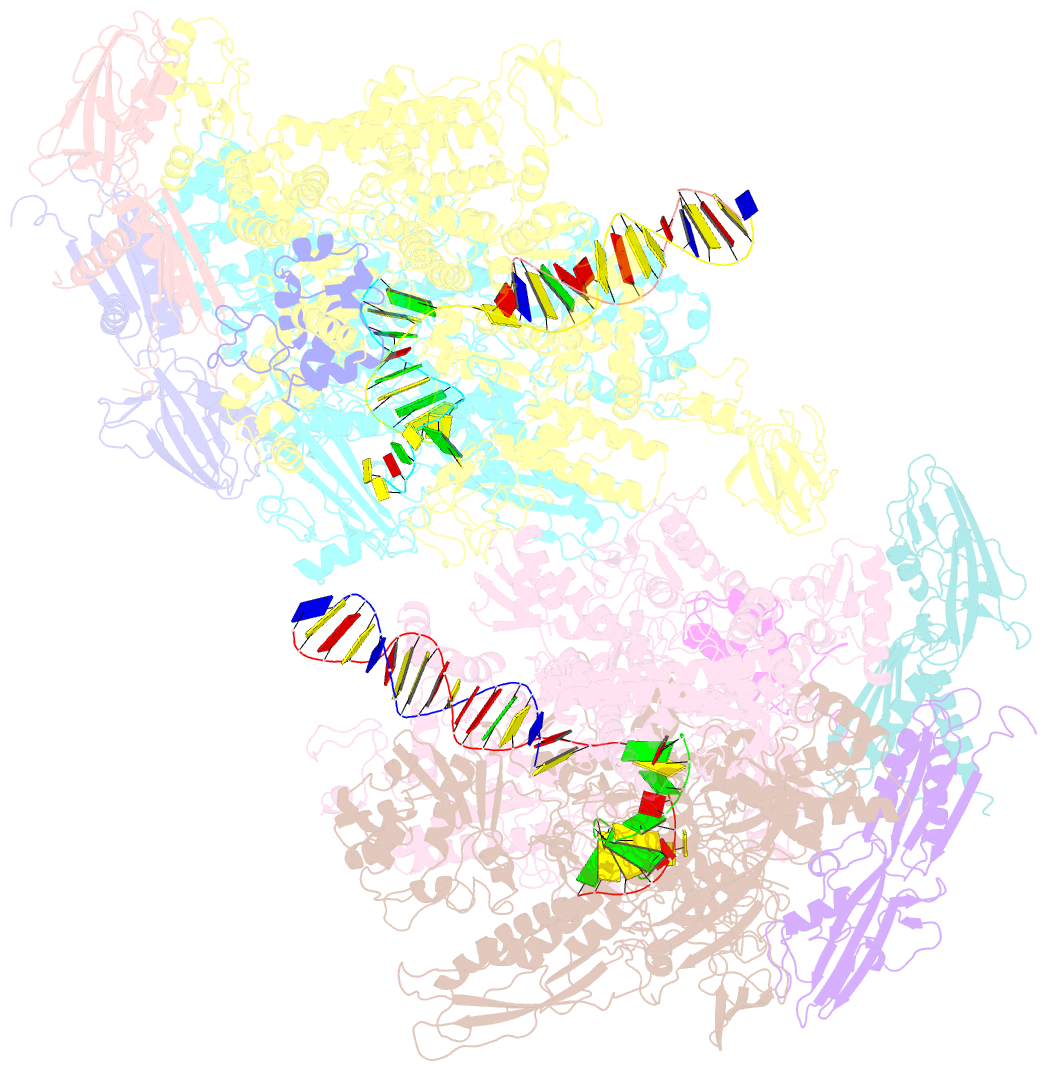
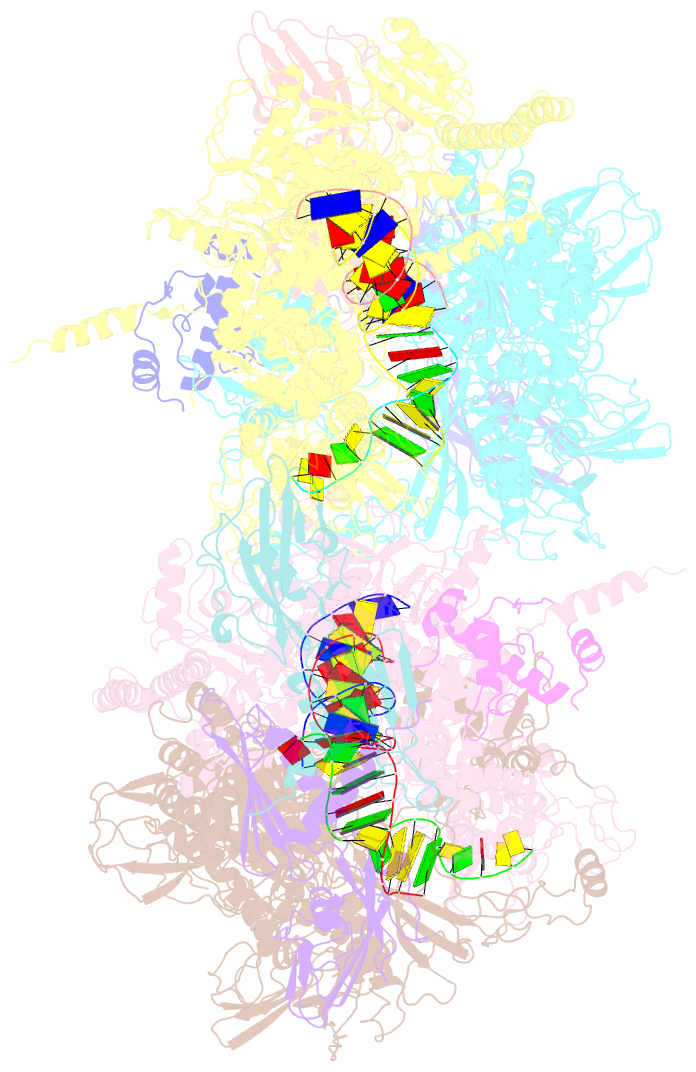
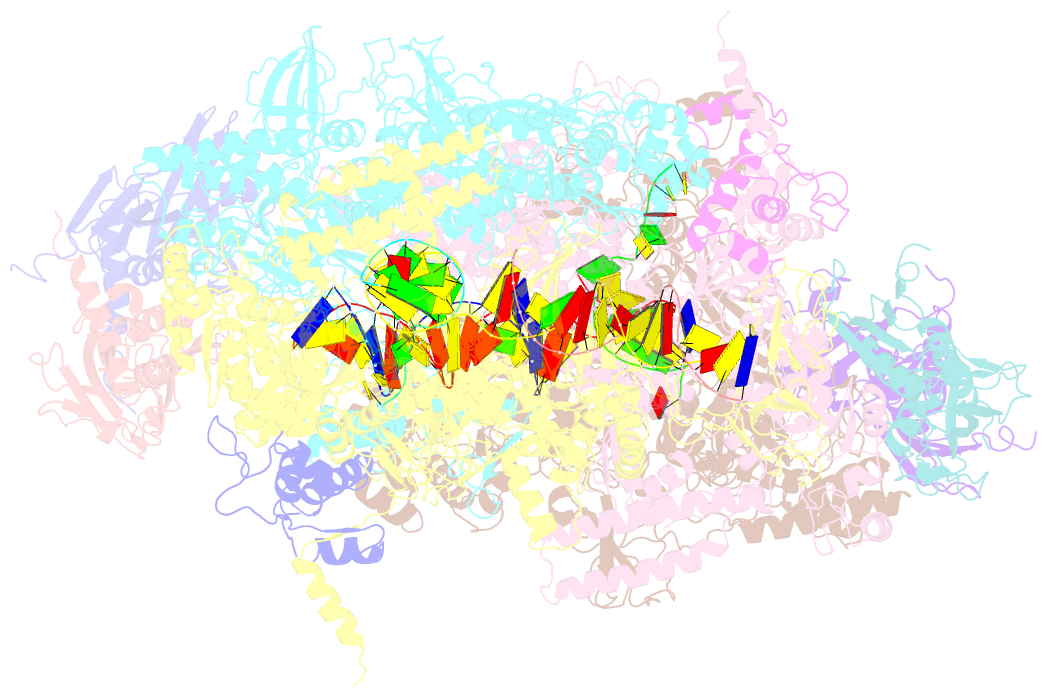
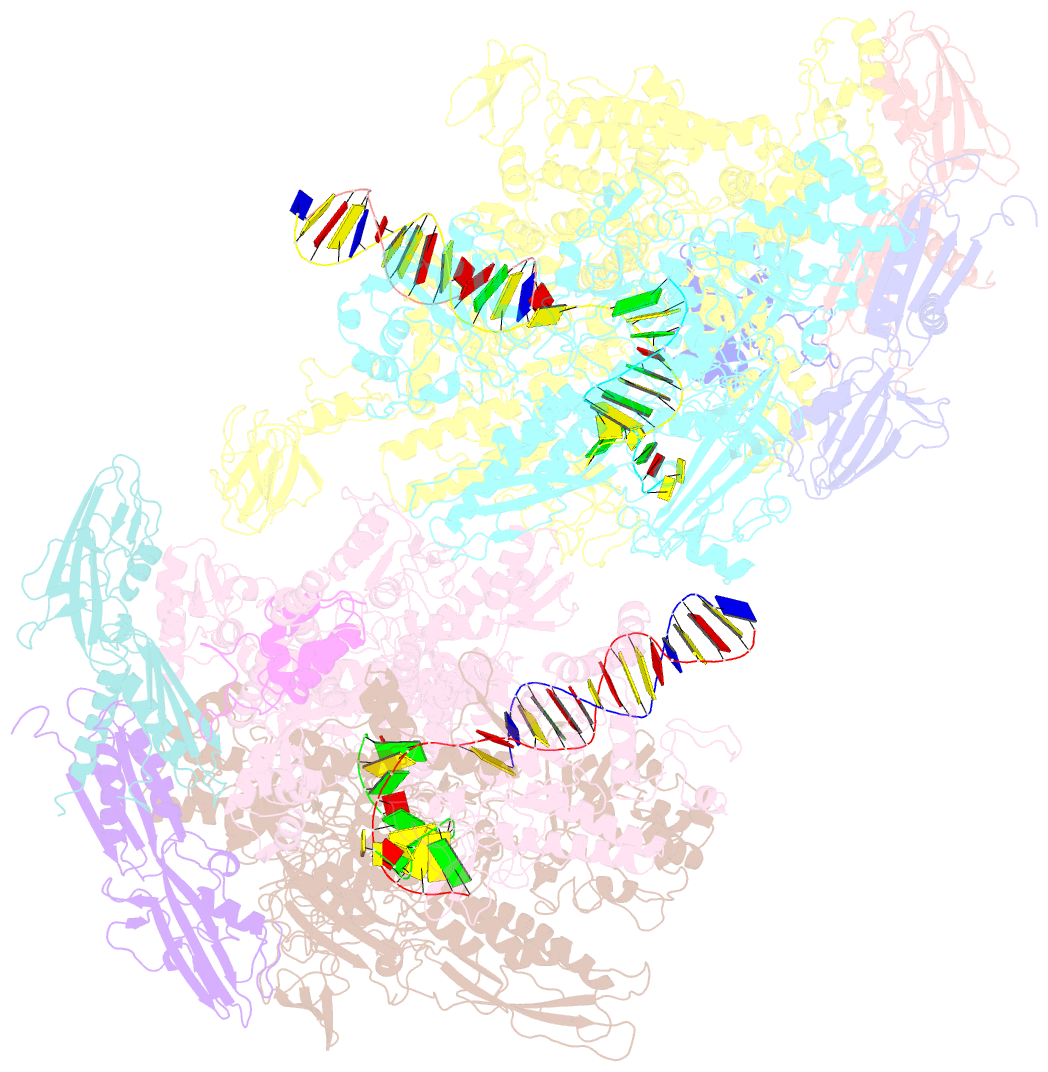
- Thermus thermophilus RNA polymerase backtracked complex
- Reference
- Sekine SI, Murayama Y, Svetlov V, Nudler E, Yokoyama S (2015): "The Ratcheted and Ratchetable Structural States of RNA Polymerase Underlie Multiple Transcriptional Functions." Mol.Cell, 57, 408-421. doi: 10.1016/j.molcel.2014.12.014.
- Abstract
- DNA-dependent RNA polymerase (RNAP) accomplishes multiple tasks during transcription by assuming different structural forms. Reportedly, the "tight" form performs nucleotide addition to nascent RNA, while the "ratcheted" form is adopted for transcription inhibition. In this study, we performed Cys-pair crosslinking (CPX) analyses of various transcription complexes of a bacterial RNAP and crystallographic analyses of its backtracked and Gre-factor-bound states to clarify which of the two forms is adopted. The ratcheted form was revealed to support GreA-dependent transcript cleavage, long backtracking, hairpin-dependent pausing, and termination. In contrast, the tight form correlated with nucleotide addition, mismatch-dependent pausing, one-nucleotide backtracking, and factor-independent transcript cleavage. RNAP in the paused/backtracked state, but not the nucleotide-addition state, readily transitions to the ratcheted form ("ratchetable"), indicating that the tight form represents two distinct regulatory states. The 3' end and the hairpin structure of the nascent RNA promote the ratchetable nature by modulating the trigger-loop conformation.