Summary information and primary citation
- PDB-id
- 4wrt; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-RNA
- Method
- X-ray (2.7 Å)
- Summary
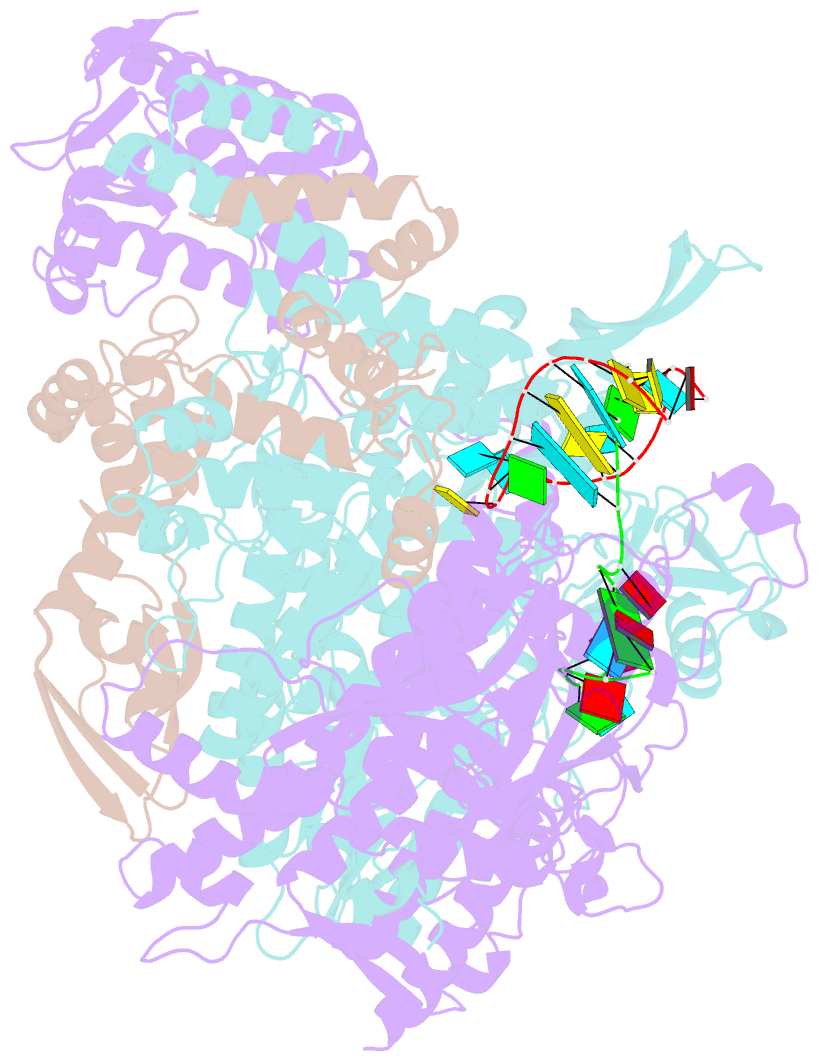
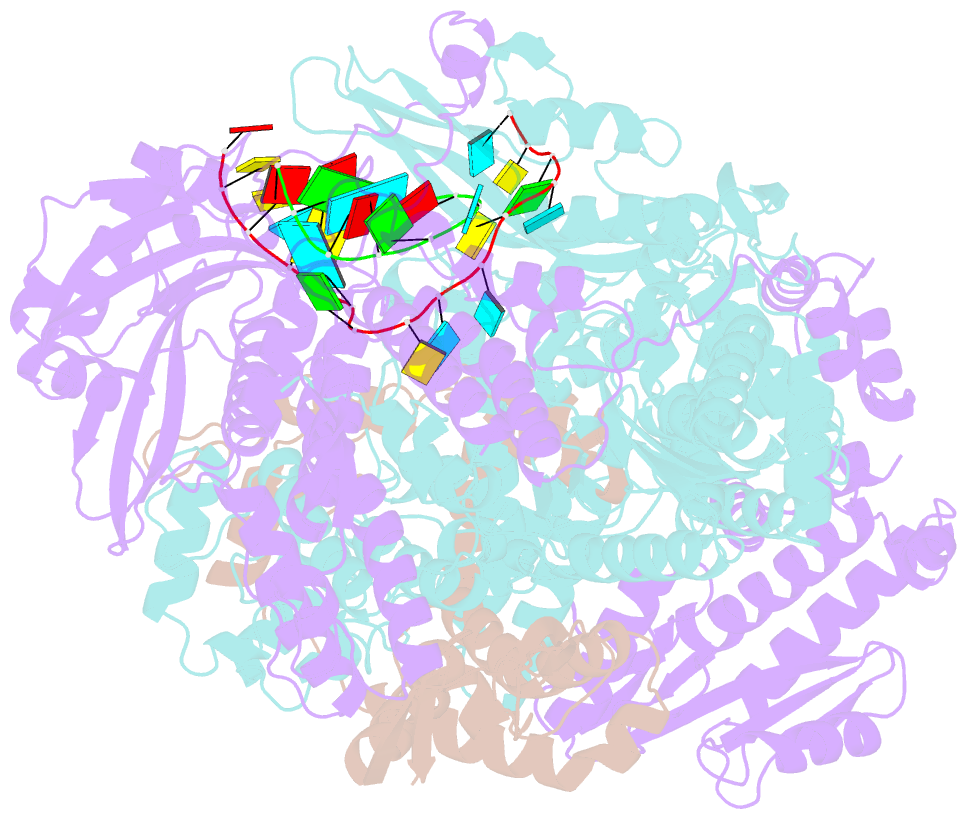
- Crystal structure of influenza b polymerase with bound vrna promoter (form flub2)
- Reference
- Reich S, Guilligay D, Pflug A, Malet H, Berger I, Crepin T, Hart D, Lunardi T, Nanao M, Ruigrok RW, Cusack S (2014): "Structural insight into cap-snatching and RNA synthesis by influenza polymerase." Nature, 516, 361-366. doi: 10.1038/nature14009.
- Abstract
- Influenza virus polymerase uses a capped primer, derived by 'cap-snatching' from host pre-messenger RNA, to transcribe its RNA genome into mRNA and a stuttering mechanism to generate the poly(A) tail. By contrast, genome replication is unprimed and generates exact full-length copies of the template. Here we use crystal structures of bat influenza A and human influenza B polymerases (FluA and FluB), bound to the viral RNA promoter, to give mechanistic insight into these distinct processes. In the FluA structure, a loop analogous to the priming loop of flavivirus polymerases suggests that influenza could initiate unprimed template replication by a similar mechanism. Comparing the FluA and FluB structures suggests that cap-snatching involves in situ rotation of the PB2 cap-binding domain to direct the capped primer first towards the endonuclease and then into the polymerase active site. The polymerase probably undergoes considerable conformational changes to convert the observed pre-initiation state into the active initiation and elongation states.