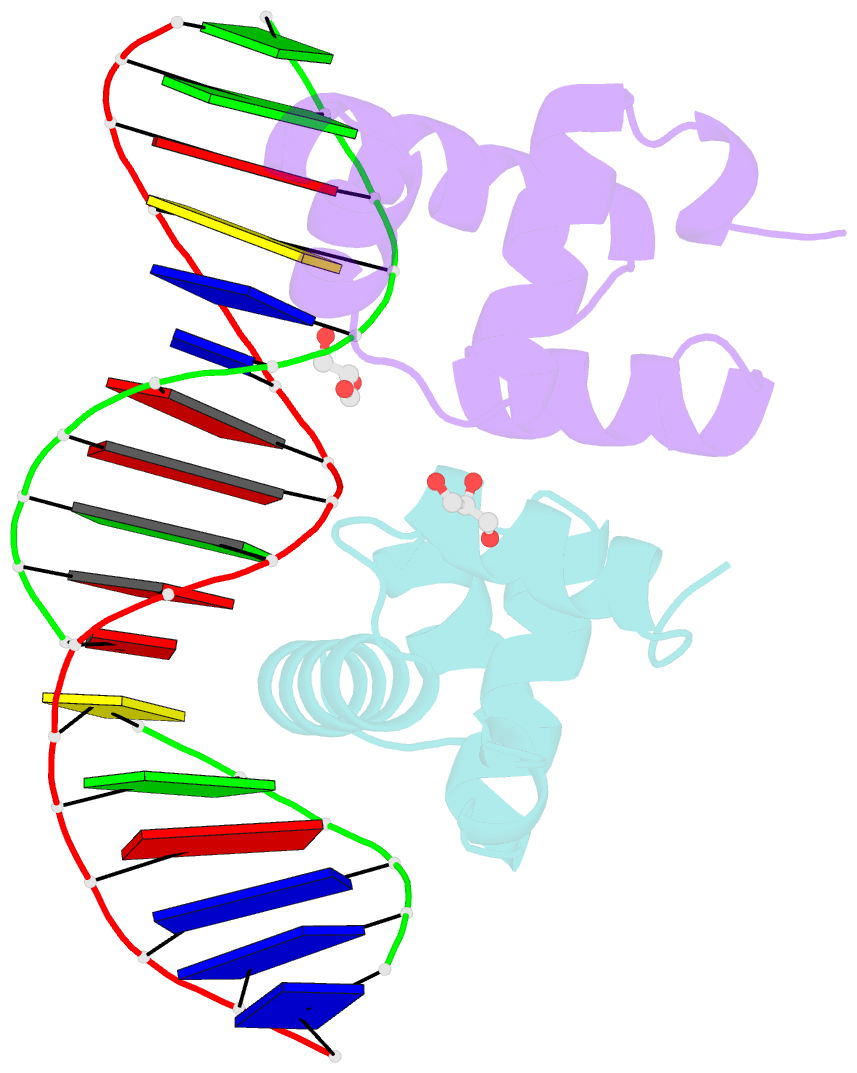
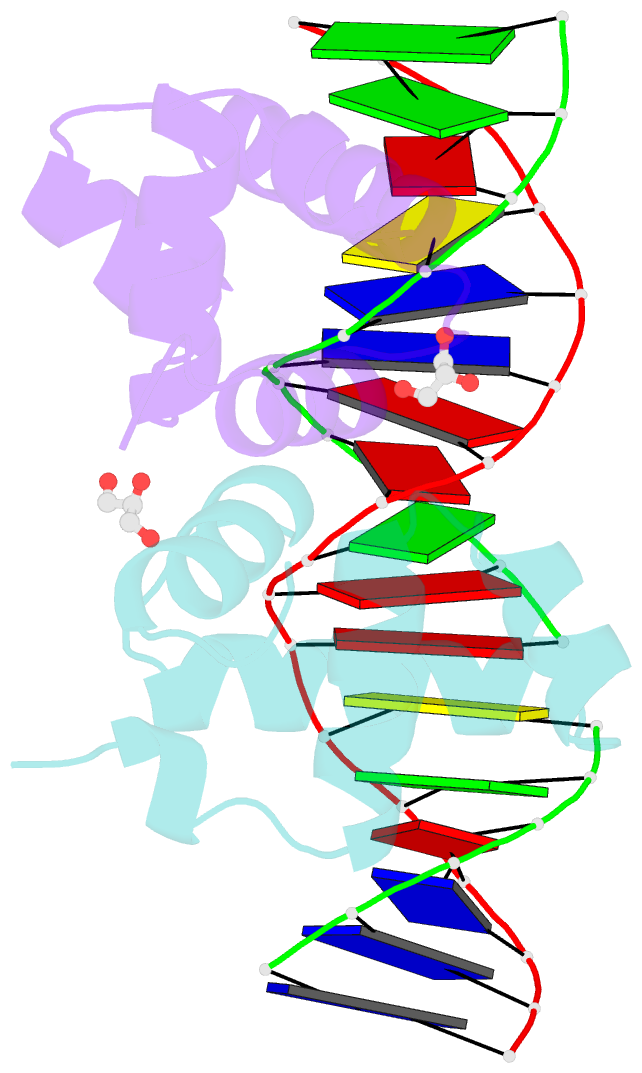
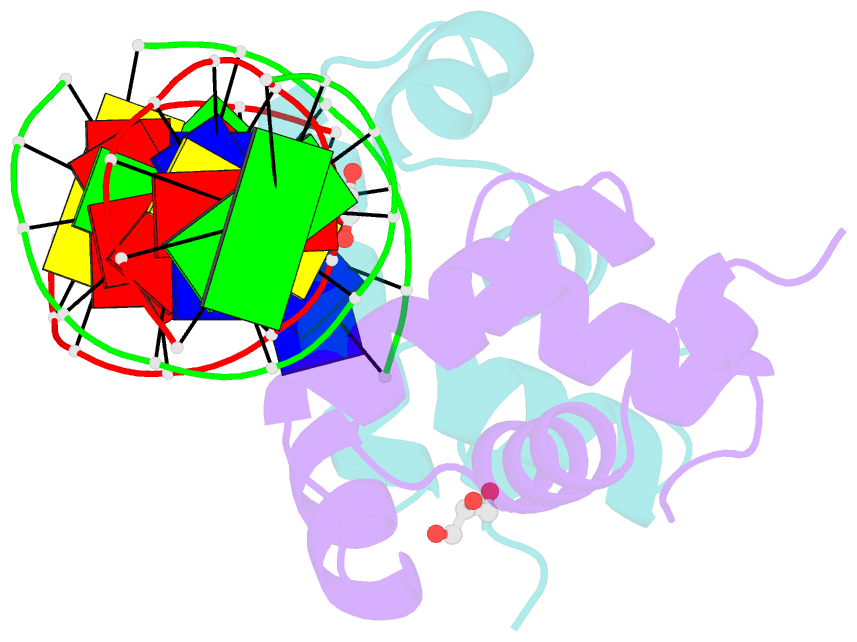
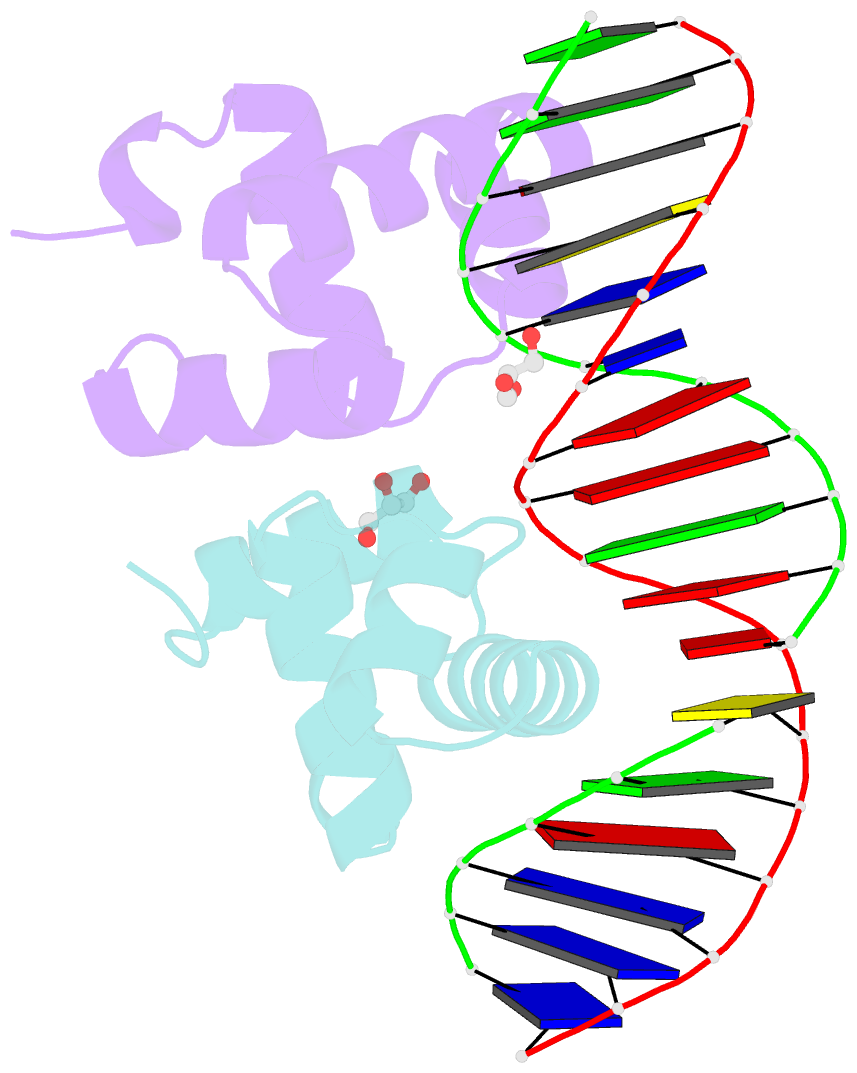
Summary information and primary citation
- PDB-id
- 4wu4; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.3 Å)
- Summary
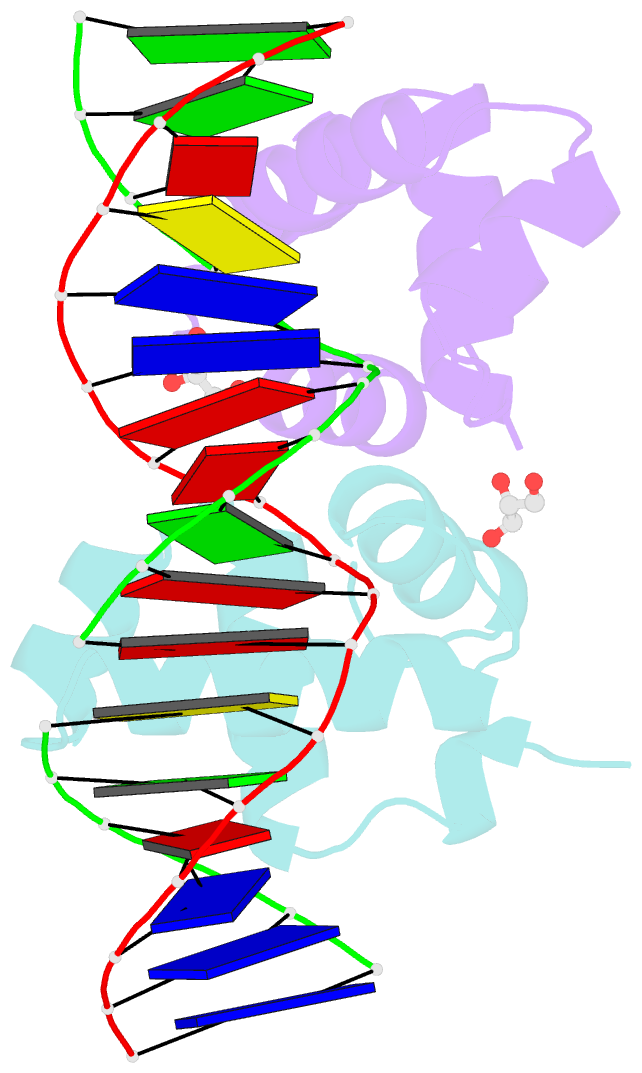
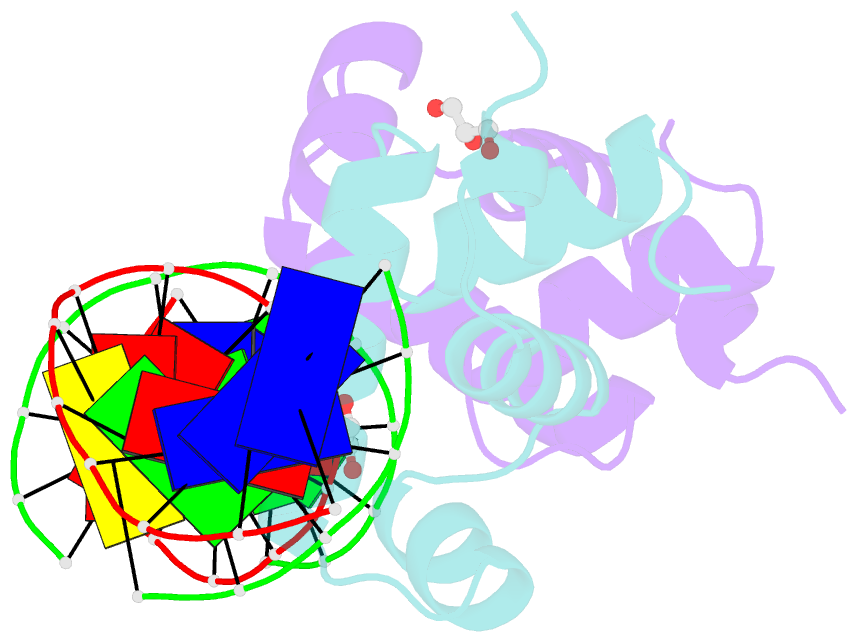
- Crystal structure of e. faecalis DNA binding domain liard191n complexed with 22bp DNA
- Reference
- Davlieva M, Shi Y, Leonard PG, Johnson TA, Zianni MR, Arias CA, Ladbury JE, Shamoo Y (2015): "A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin." Nucleic Acids Res., 43, 4758-4773. doi: 10.1093/nar/gkv321.
- Abstract
- LiaR is a 'master regulator' of the cell envelope stress response in enterococci and many other Gram-positive organisms. Mutations to liaR can lead to antibiotic resistance to a variety of antibiotics including the cyclic lipopeptide daptomycin. LiaR is phosphorylated in response to membrane stress to regulate downstream target operons. Using DNA footprinting of the regions upstream of the liaXYZ and liaFSR operons we show that LiaR binds an extended stretch of DNA that extends beyond the proposed canonical consensus sequence suggesting a more complex level of regulatory control of target operons. We go on to determine the biochemical and structural basis for increased resistance to daptomycin by the adaptive mutation to LiaR (D191N) first identified from the pathogen Enterococcus faecalis S613. LiaR(D191N) increases oligomerization of LiaR to form a constitutively activated tetramer that has high affinity for DNA even in the absence of phosphorylation leading to increased resistance. Crystal structures of the LiaR DNA binding domain complexed to the putative consensus sequence as well as an adjoining secondary sequence show that upon binding, LiaR induces DNA bending that is consistent with increased recruitment of RNA polymerase to the transcription start site and upregulation of target operons.