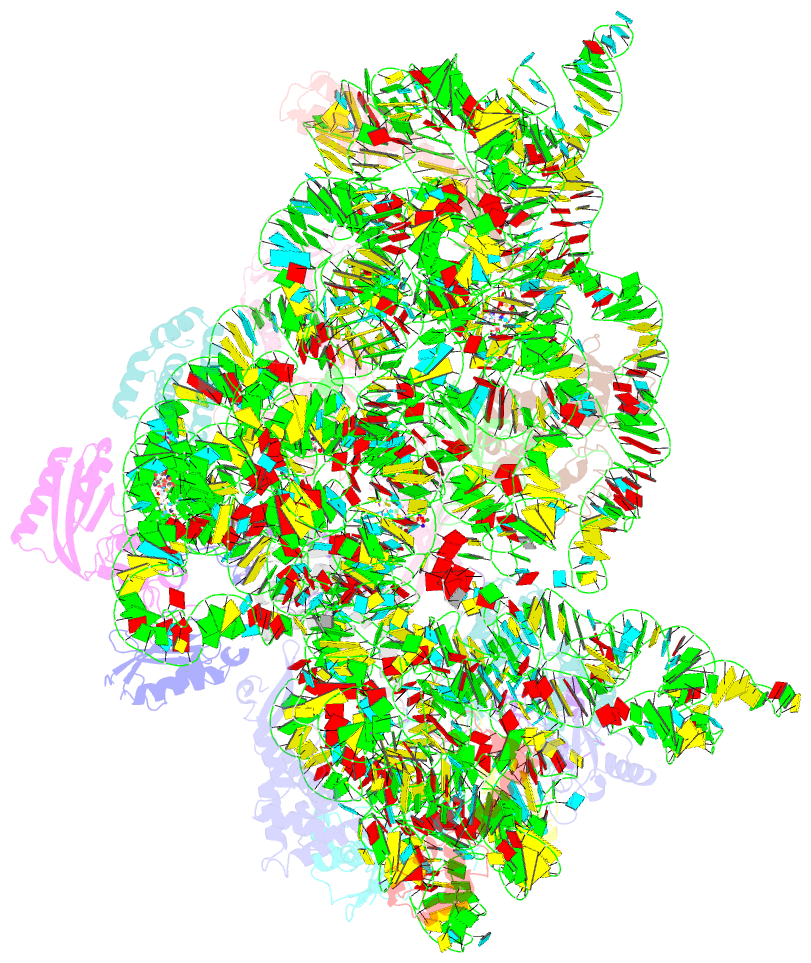
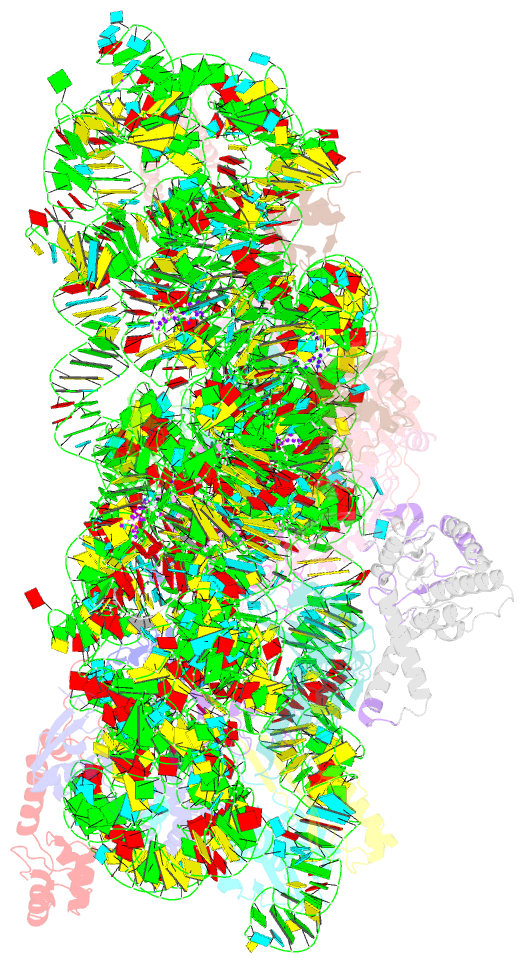
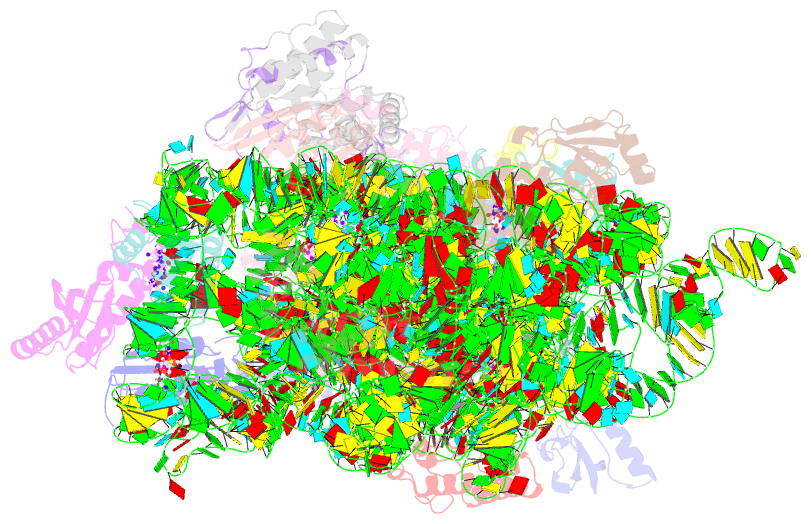
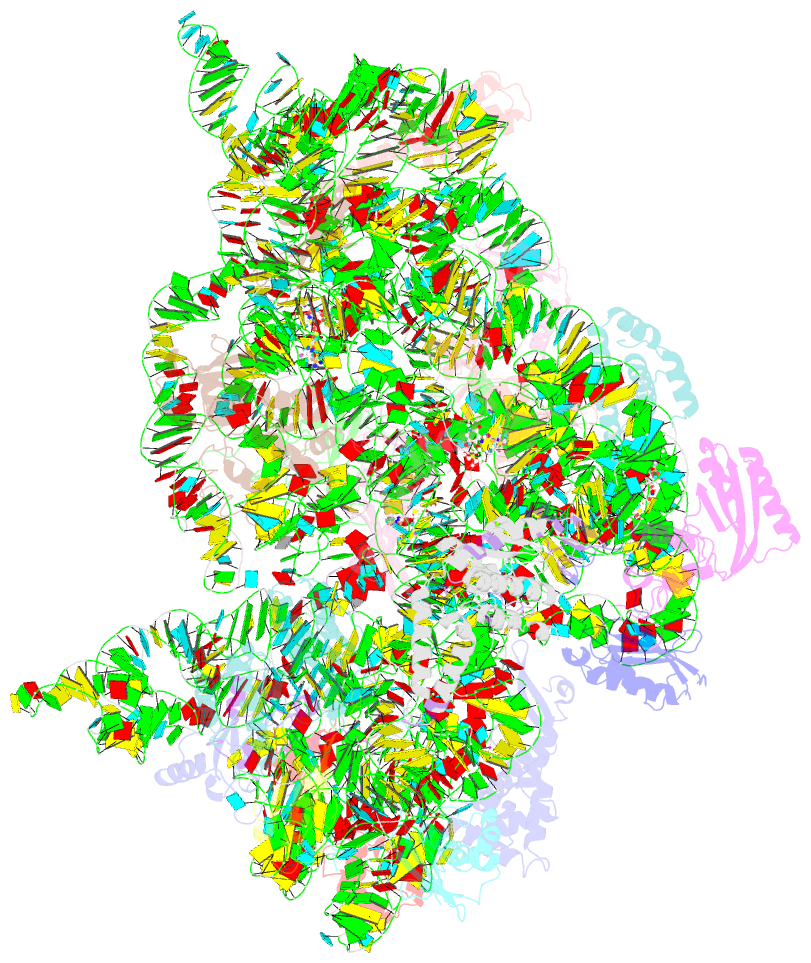
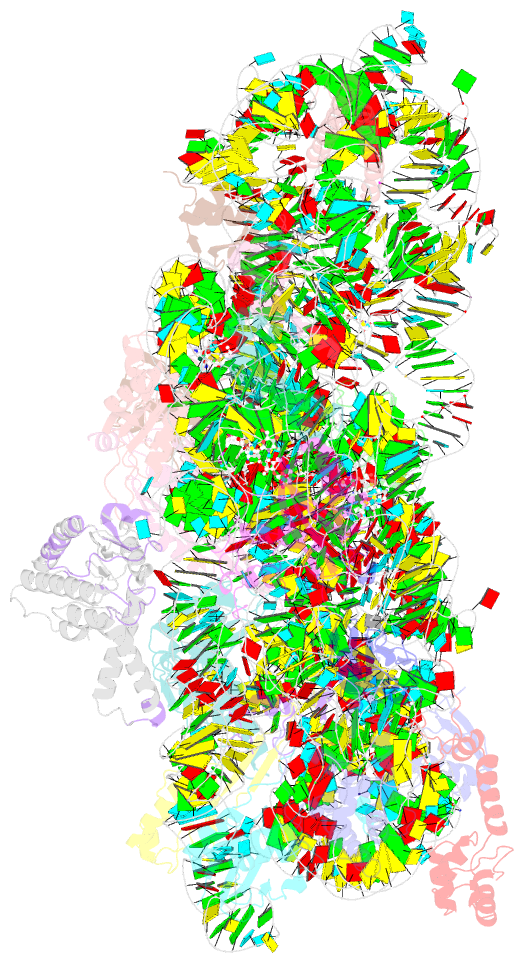
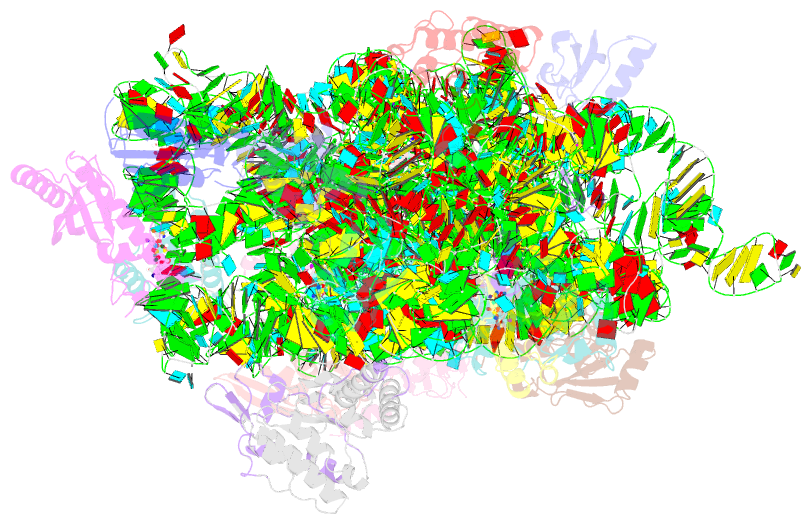
Summary information and primary citation
- PDB-id
- 4x66; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome-antibiotic
- Method
- X-ray (3.446 Å)
- Summary
- Crystal structure of 30s ribosomal subunit from thermus thermophilus
- Reference
- Choi J, Ieong KW, Demirci H, Chen J, Petrov A, Prabhakar A, O'Leary SE, Dominissini D, Rechavi G, Soltis SM, Ehrenberg M, Puglisi JD (2016): "N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics." Nat.Struct.Mol.Biol., 23, 110-115. doi: 10.1038/nsmb.3148.
- Abstract
- N(6)-methylation of adenosine (forming m(6)A) is the most abundant post-transcriptional modification within the coding region of mRNA, but its role during translation remains unknown. Here, we used bulk kinetic and single-molecule methods to probe the effect of m(6)A in mRNA decoding. Although m(6)A base-pairs with uridine during decoding, as shown by X-ray crystallographic analyses of Thermus thermophilus ribosomal complexes, our measurements in an Escherichia coli translation system revealed that m(6)A modification of mRNA acts as a barrier to tRNA accommodation and translation elongation. The interaction between an m(6)A-modified codon and cognate tRNA echoes the interaction between a near-cognate codon and tRNA, because delay in tRNA accommodation depends on the position and context of m(6)A within codons and on the accuracy level of translation. Overall, our results demonstrate that chemical modification of mRNA can change translational dynamics.