Summary information and primary citation
- PDB-id
- 4x9e; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (3.1 Å)
- Summary
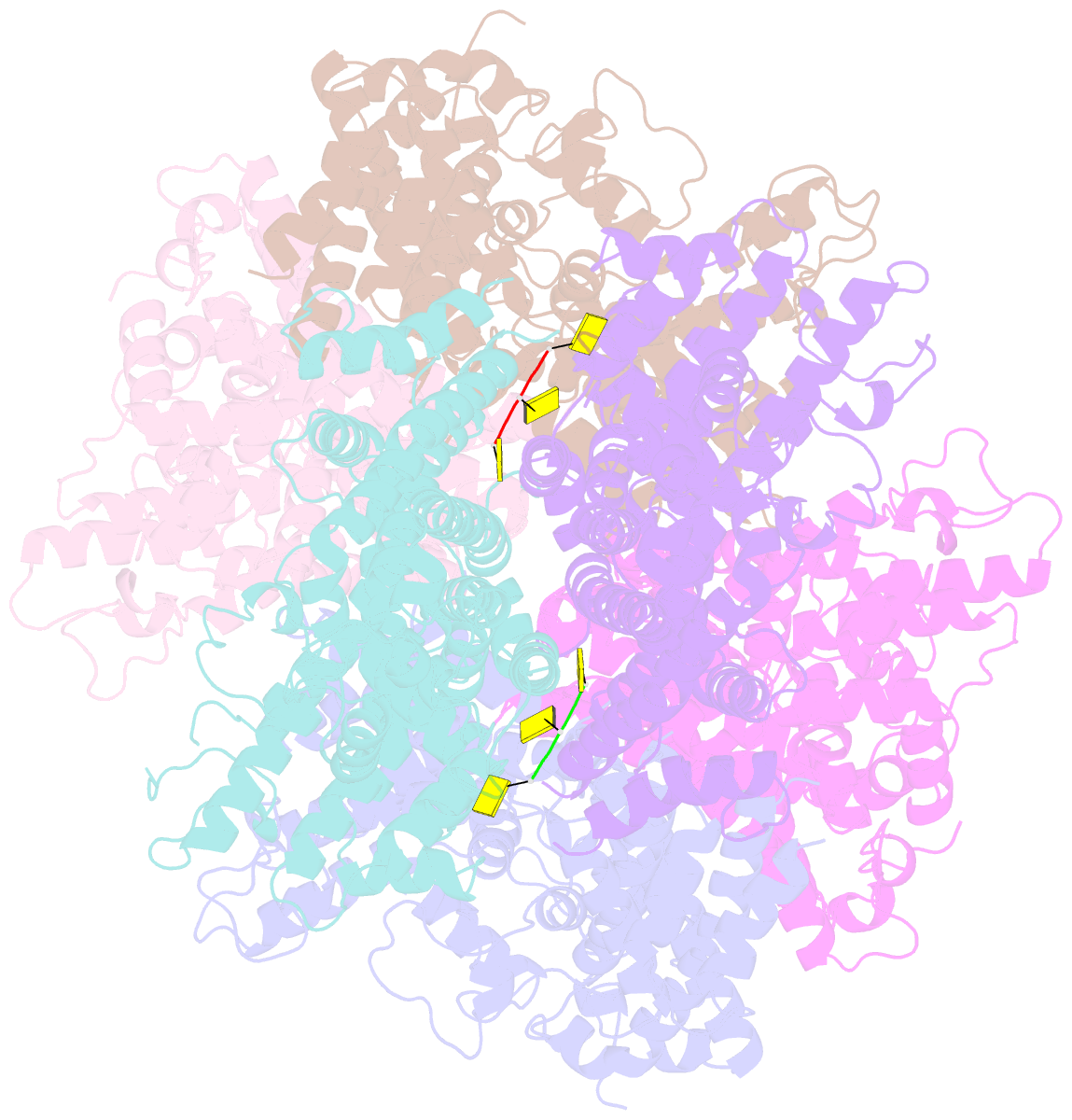
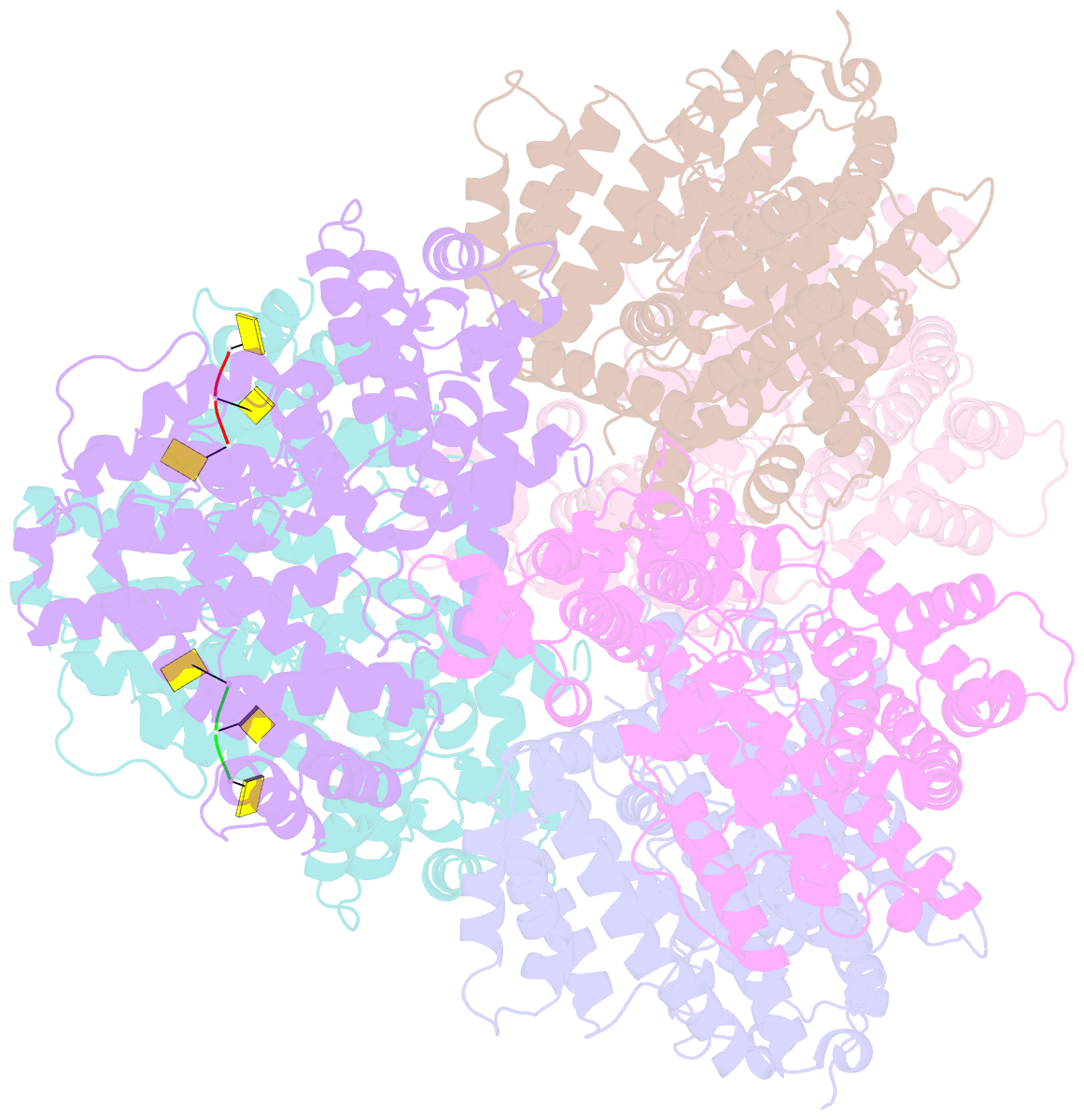
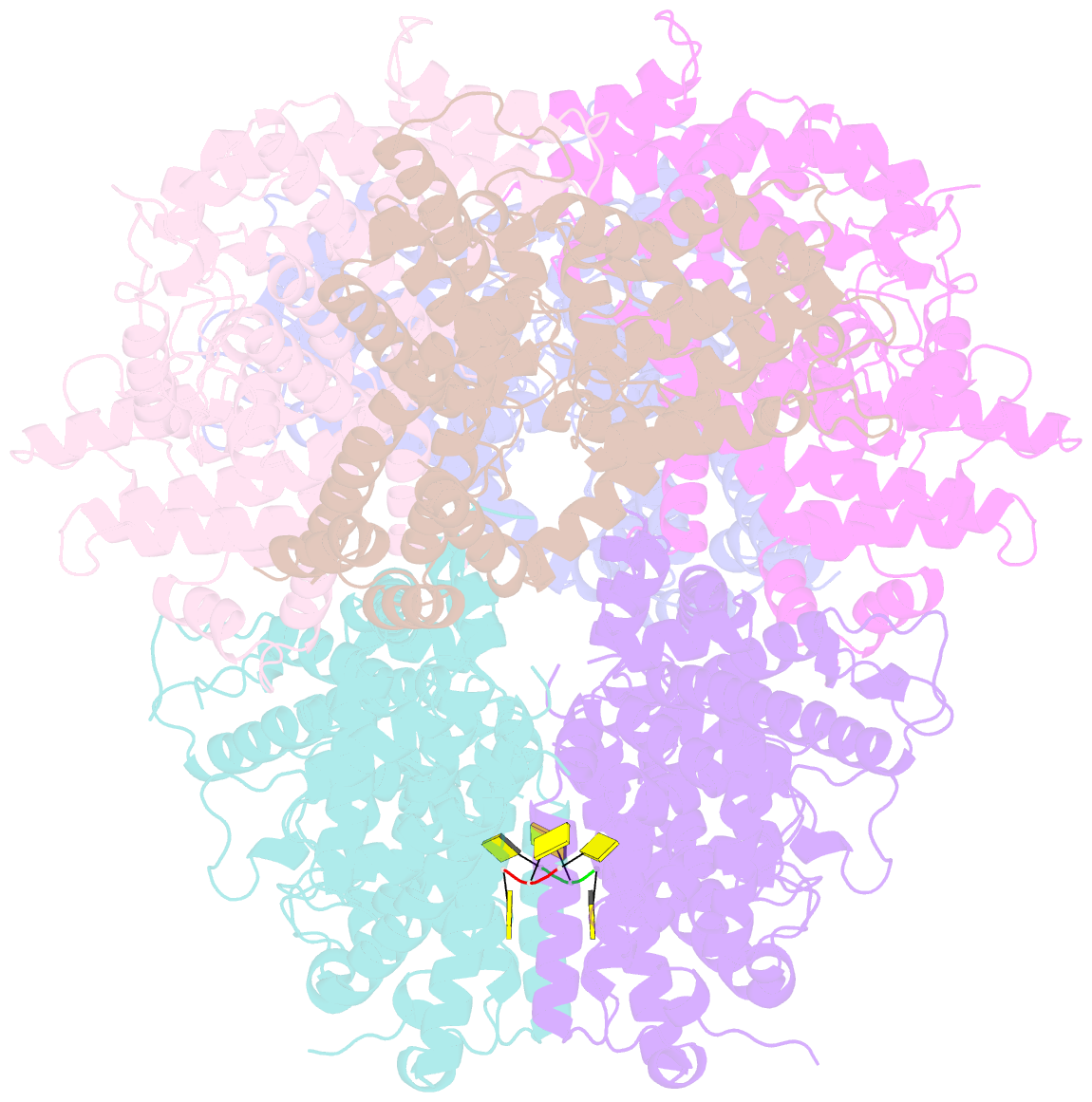
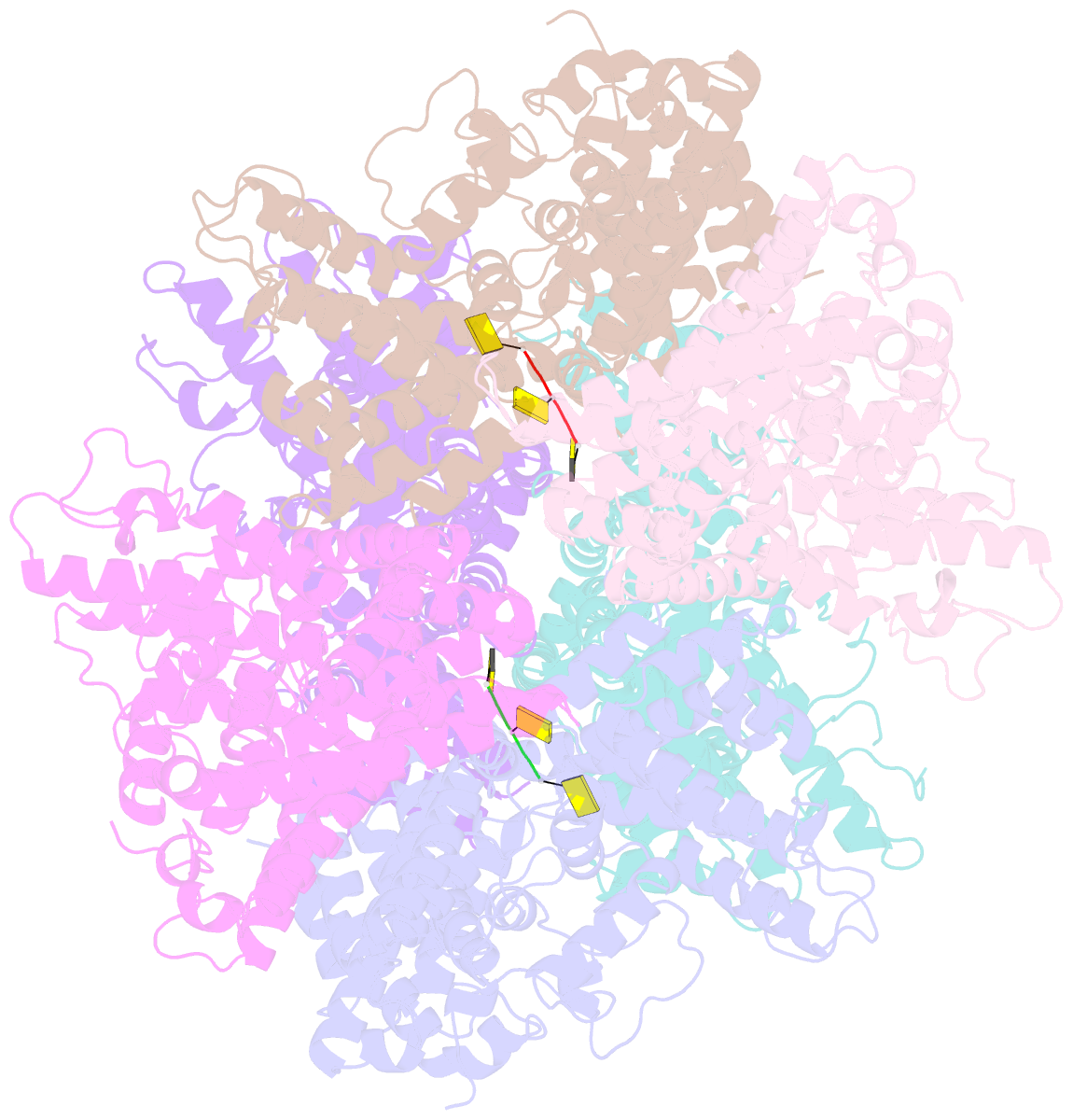
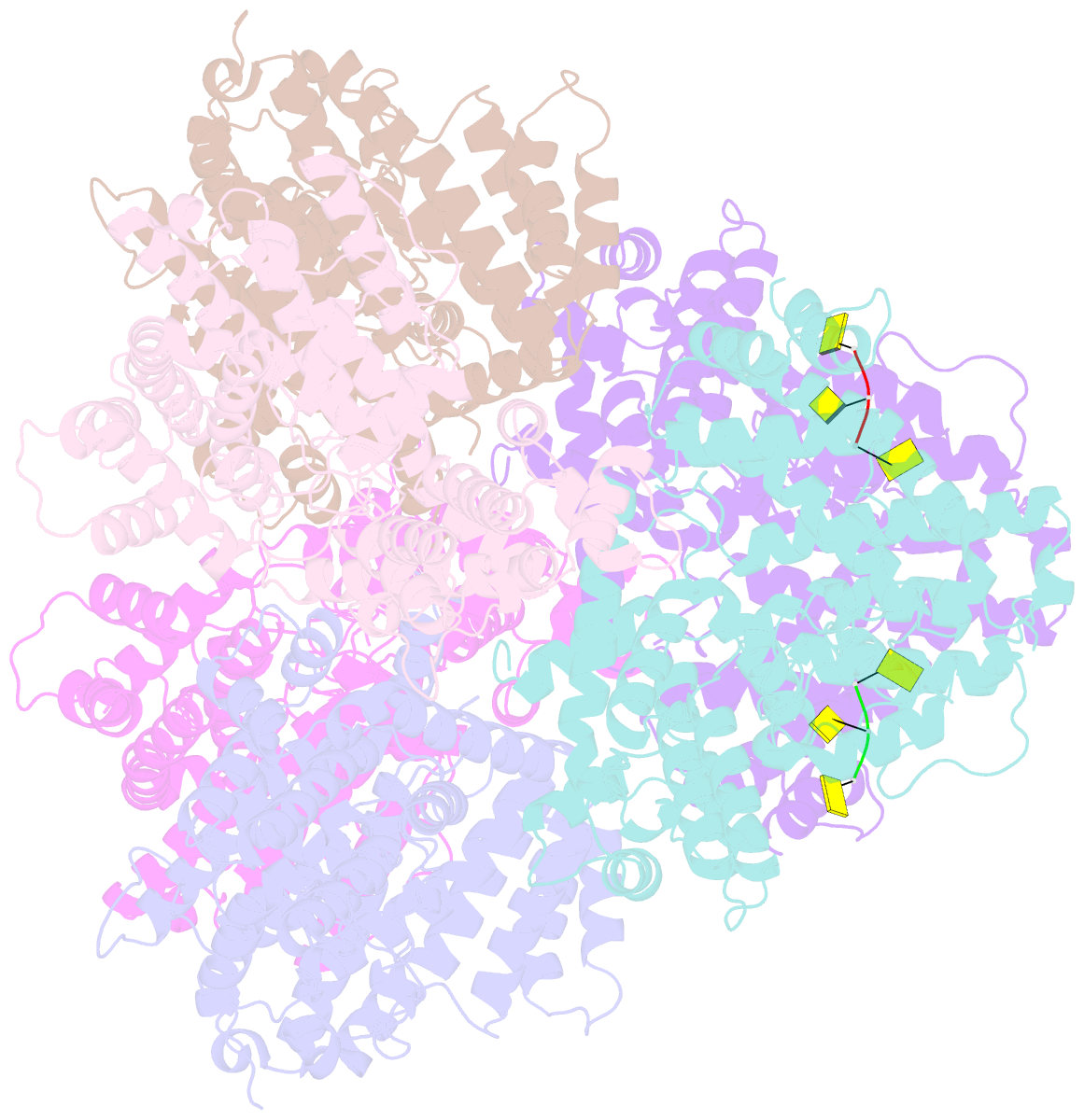
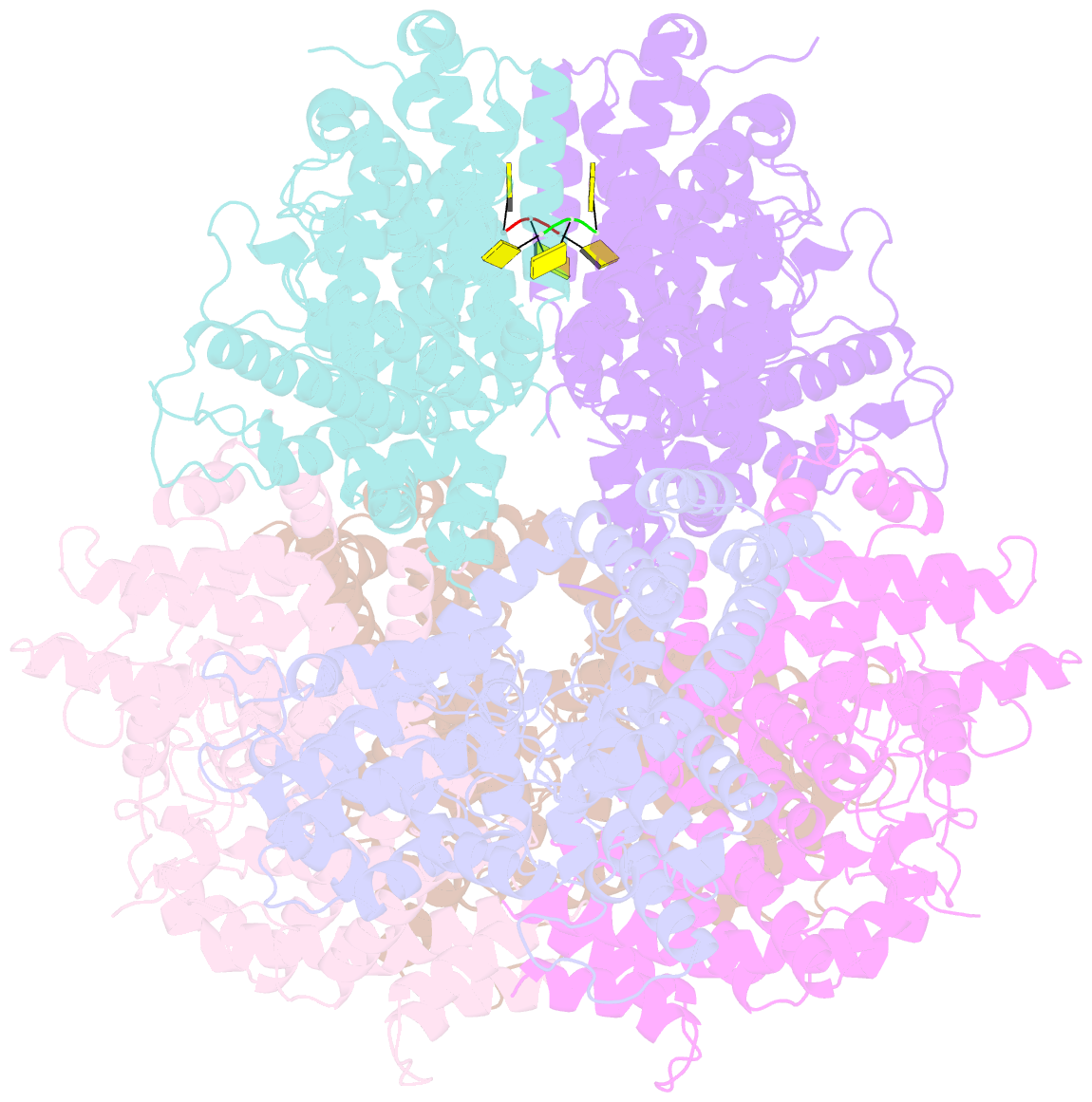
- Deoxyguanosinetriphosphate triphosphohydrolase from escherichia coli with two DNA effector molecules
- Reference
- Singh D, Gawel D, Itsko M, Hochkoeppler A, Krahn JM, London RE, Schaaper RM (2015): "Structure of Escherichia coli dGTP Triphosphohydrolase: A HEXAMERIC ENZYME WITH DNA EFFECTOR MOLECULES." J.Biol.Chem., 290, 10418-10429. doi: 10.1074/jbc.M115.636936.
- Abstract
- The Escherichia coli dgt gene encodes a dGTP triphosphohydrolase whose detailed role still remains to be determined. Deletion of dgt creates a mutator phenotype, indicating that the dGTPase has a fidelity role, possibly by affecting the cellular dNTP pool. In the present study, we have investigated the structure of the Dgt protein at 3.1-Å resolution. One of the obtained structures revealed a protein hexamer that contained two molecules of single-stranded DNA. The presence of DNA caused significant conformational changes in the enzyme, including in the catalytic site of the enzyme. Dgt preparations lacking DNA were able to bind single-stranded DNA with high affinity (Kd ∼ 50 nM). DNA binding positively affected the activity of the enzyme: dGTPase activity displayed sigmoidal (cooperative) behavior without DNA but hyperbolic (Michaelis-Menten) kinetics in its presence, consistent with a specific lowering of the apparent Km for dGTP. A mutant Dgt enzyme was also created containing residue changes in the DNA binding cleft. This mutant enzyme, whereas still active, was incapable of DNA binding and could no longer be stimulated by addition of DNA. We also created an E. coli strain containing the mutant dgt gene on the chromosome replacing the wild-type gene. The mutant also displayed a mutator phenotype. Our results provide insight into the allosteric regulation of the enzyme and support a physiologically important role of DNA binding.