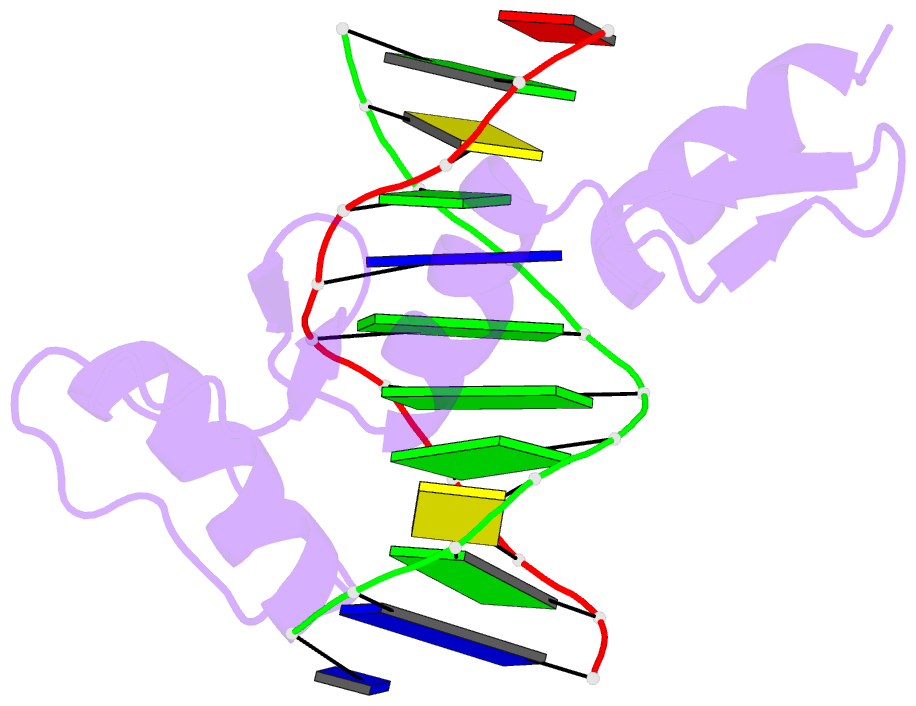
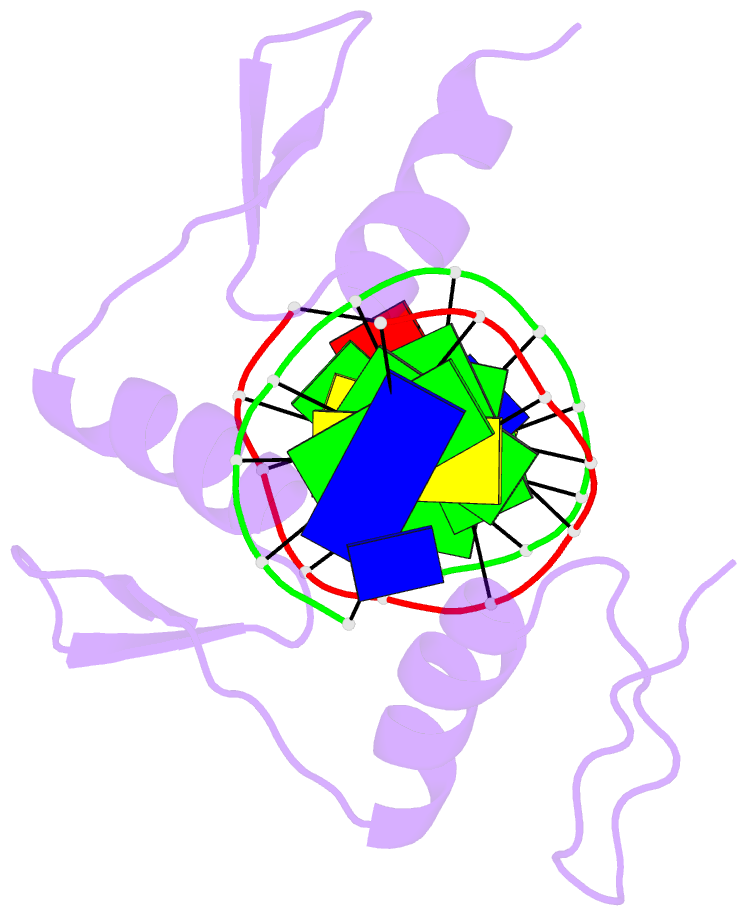
Summary information and primary citation
- PDB-id
- 4x9j; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription regulator-DNA
- Method
- X-ray (1.412 Å)
- Summary
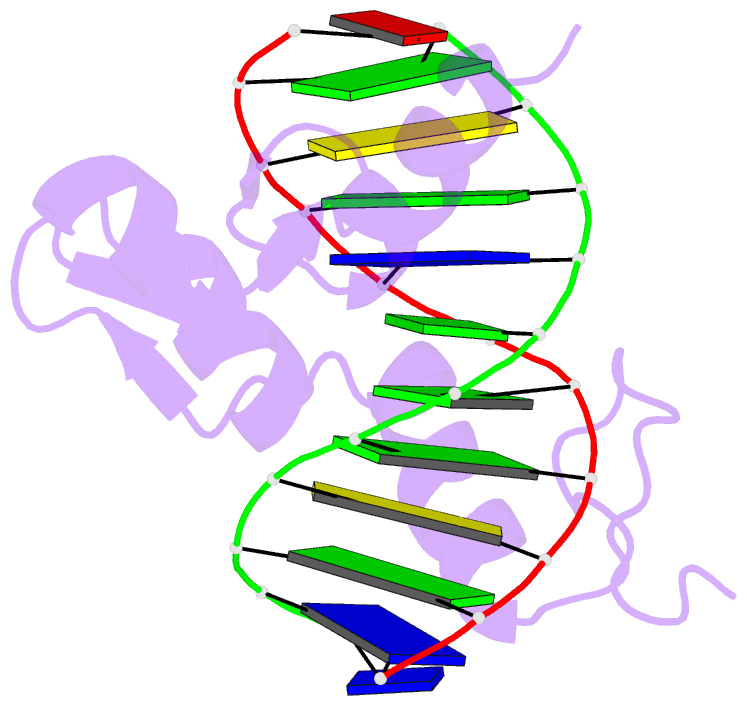
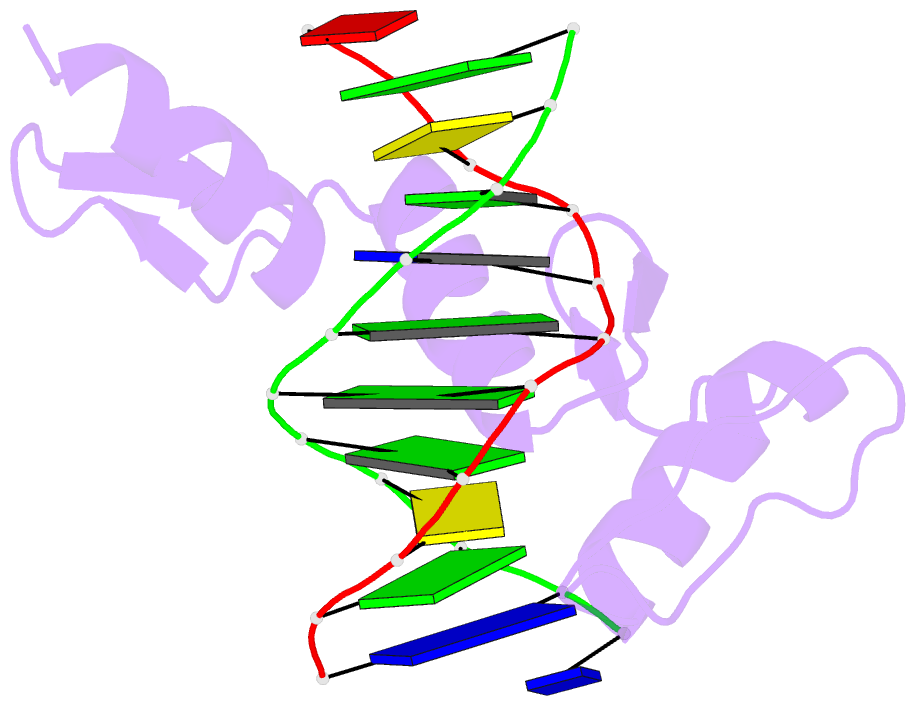
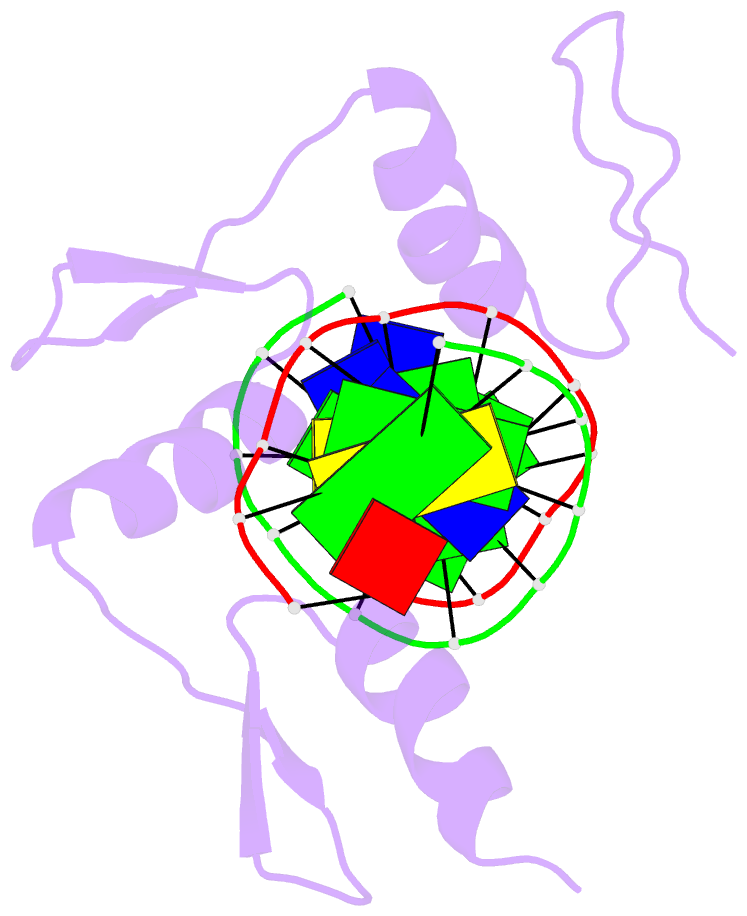
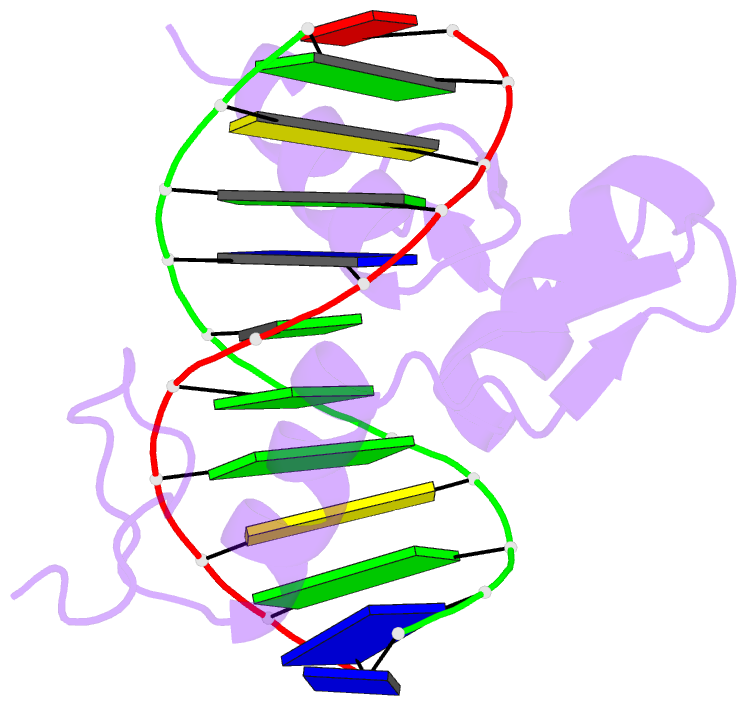
- Egr-1 with doubly methylated DNA
- Reference
- Zandarashvili L, White MA, Esadze A, Iwahara J (2015): "Structural impact of complete CpG methylation within target DNA on specific complex formation of the inducible transcription factor Egr-1." Febs Lett., 589, 1748-1753. doi: 10.1016/j.febslet.2015.05.022.
- Abstract
- The inducible transcription factor Egr-1 binds specifically to 9-bp target sequences containing two CpG sites that can potentially be methylated at four cytosine bases. Although it appears that complete CpG methylation would make an unfavorable steric clash in the previous crystal structures of the complexes with unmethylated or partially methylated DNA, our affinity data suggest that DNA recognition by Egr-1 is insensitive to CpG methylation. We have determined, at a 1.4-Å resolution, the crystal structure of the Egr-1 zinc-finger complex with completely methylated target DNA. Structural comparison of the three different methylation states reveals why Egr-1 can recognize the target sequences regardless of CpG methylation.