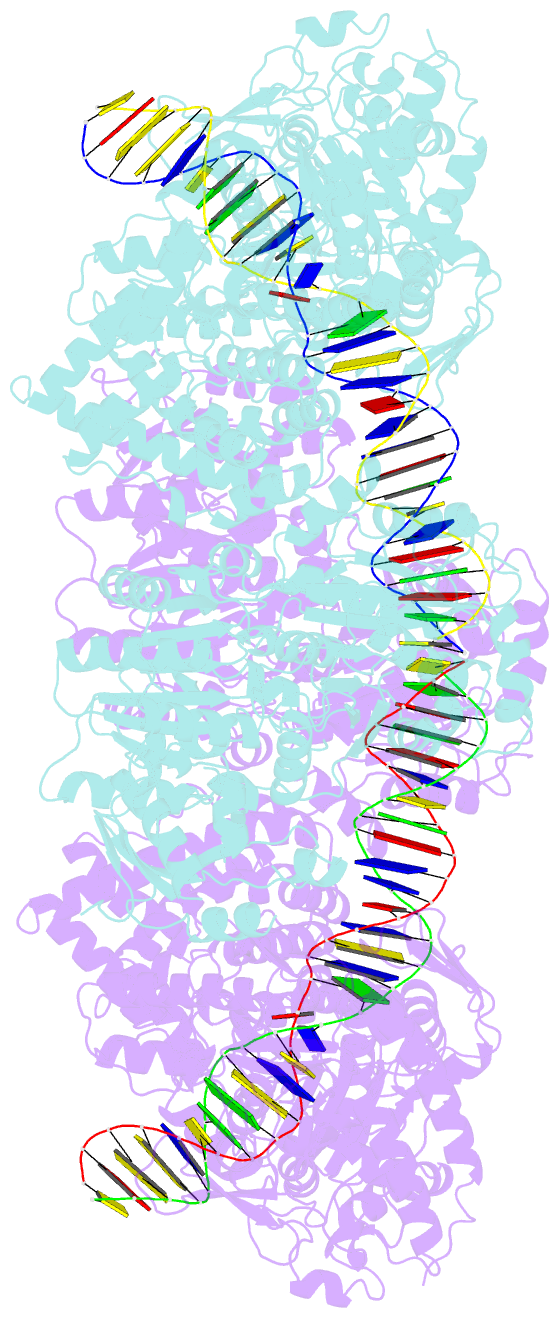
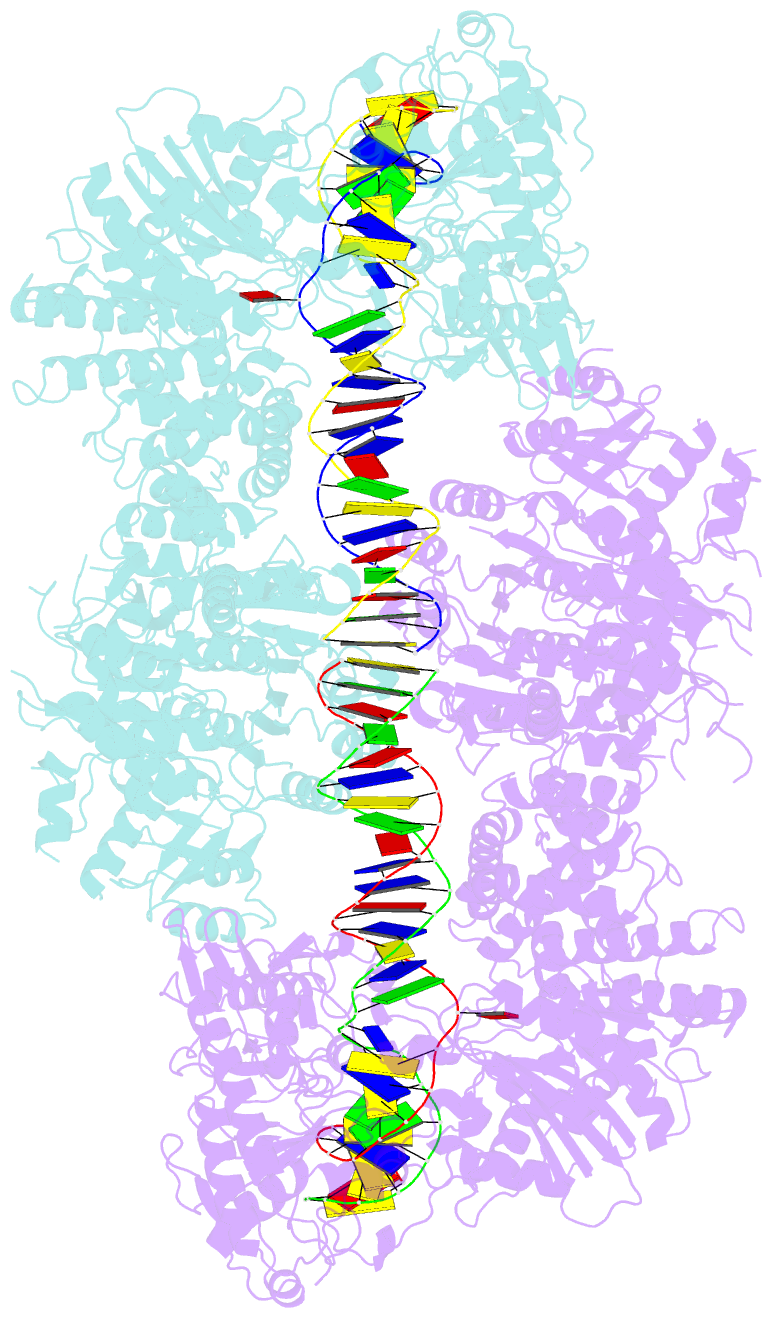
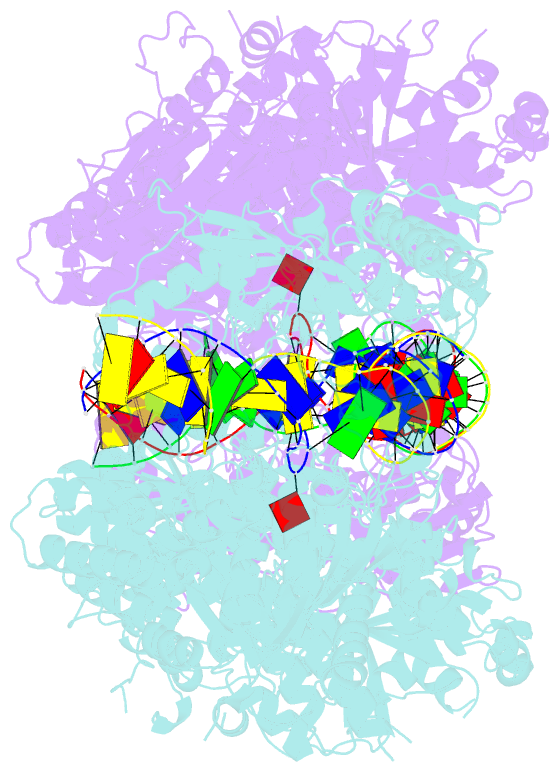
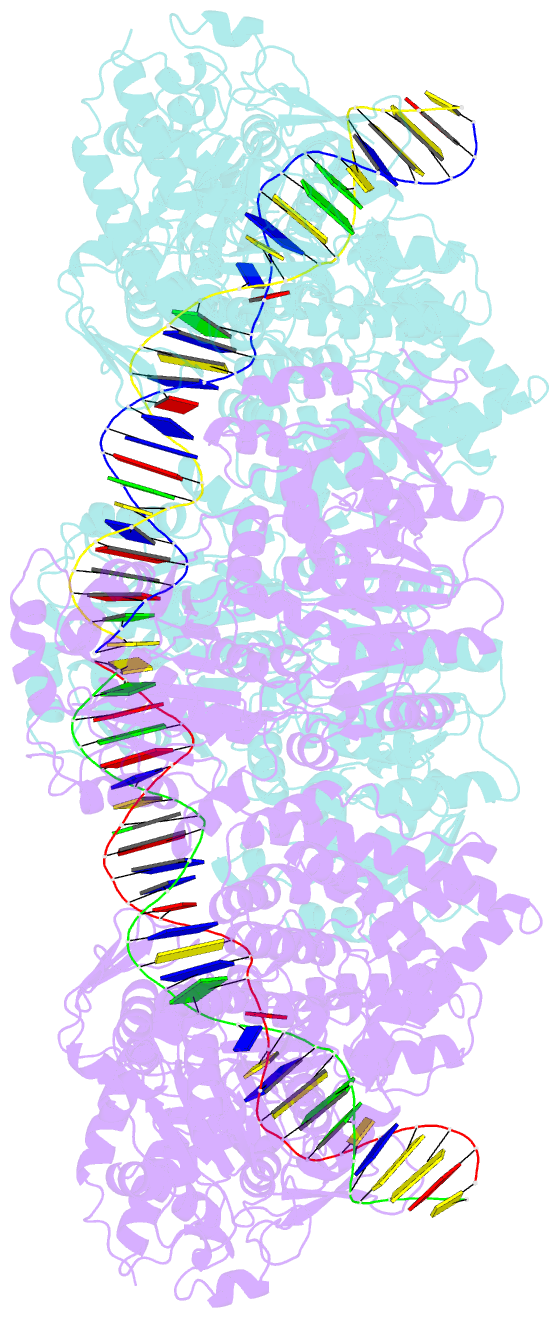
Summary information and primary citation
- PDB-id
- 4xqk; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.7 Å)
- Summary
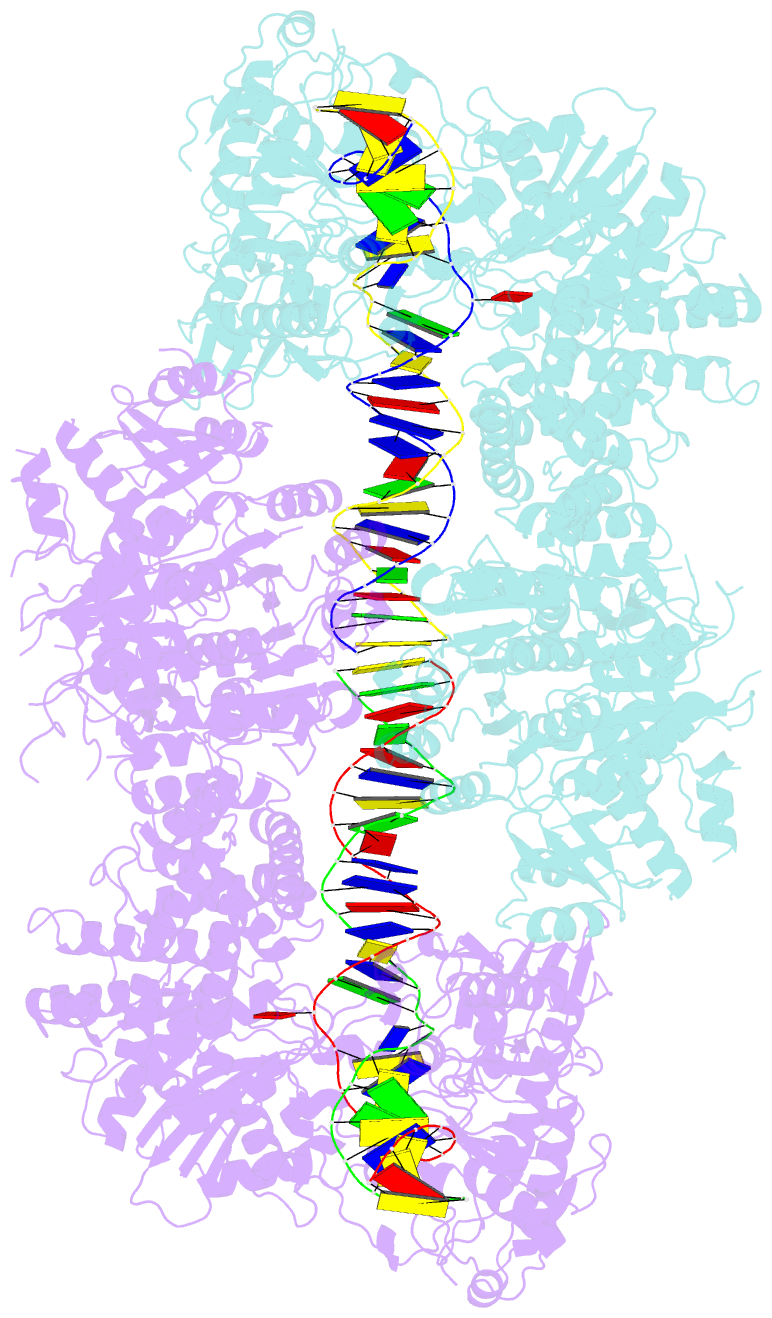
- Atp-dependent type isp restriction-modification enzyme llabiii bound to DNA
- Reference
- Chand MK, Nirwan N, Diffin FM, Aelst KV, Kulkarni M, Pernstich C, Szczelkun MD, Saikrishnan K (2015): "Translocation-coupled DNA cleavage by the Type ISP restriction-modification enzymes." Nat.Chem.Biol., 11, 870-877. doi: 10.1038/nchembio.1926.
- Abstract
- Production of endonucleolytic double-strand DNA breaks requires separate strand cleavage events. Although catalytic mechanisms for simple, dimeric endonucleases are known, there are many complex nuclease machines that are poorly understood. Here we studied the single polypeptide Type ISP restriction-modification (RM) enzymes, which cleave random DNA between distant target sites when two enzymes collide after convergent ATP-driven translocation. We report the 2.7-Å resolution X-ray crystal structure of a Type ISP enzyme-DNA complex, revealing that both the helicase-like ATPase and nuclease are located upstream of the direction of translocation, an observation inconsistent with simple nuclease-domain dimerization. Using single-molecule and biochemical techniques, we demonstrate that each ATPase remodels its DNA-protein complex and translocates along DNA without looping it, leading to a collision complex in which the nuclease domains are distal. Sequencing of the products of single cleavage events suggests a previously undescribed endonuclease model, where multiple, stochastic strand-nicking events combine to produce DNA scission.