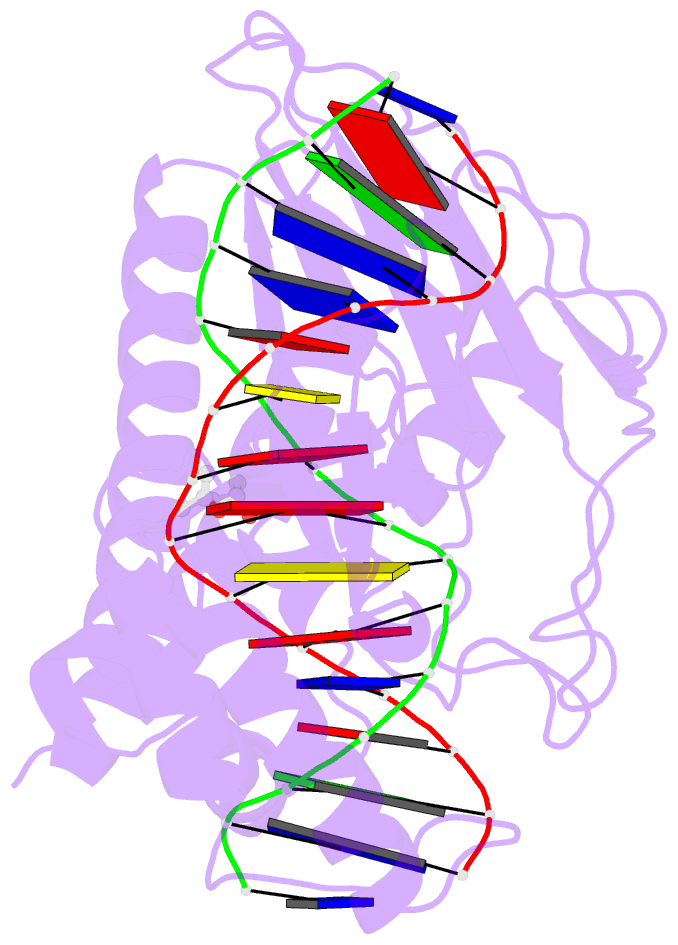
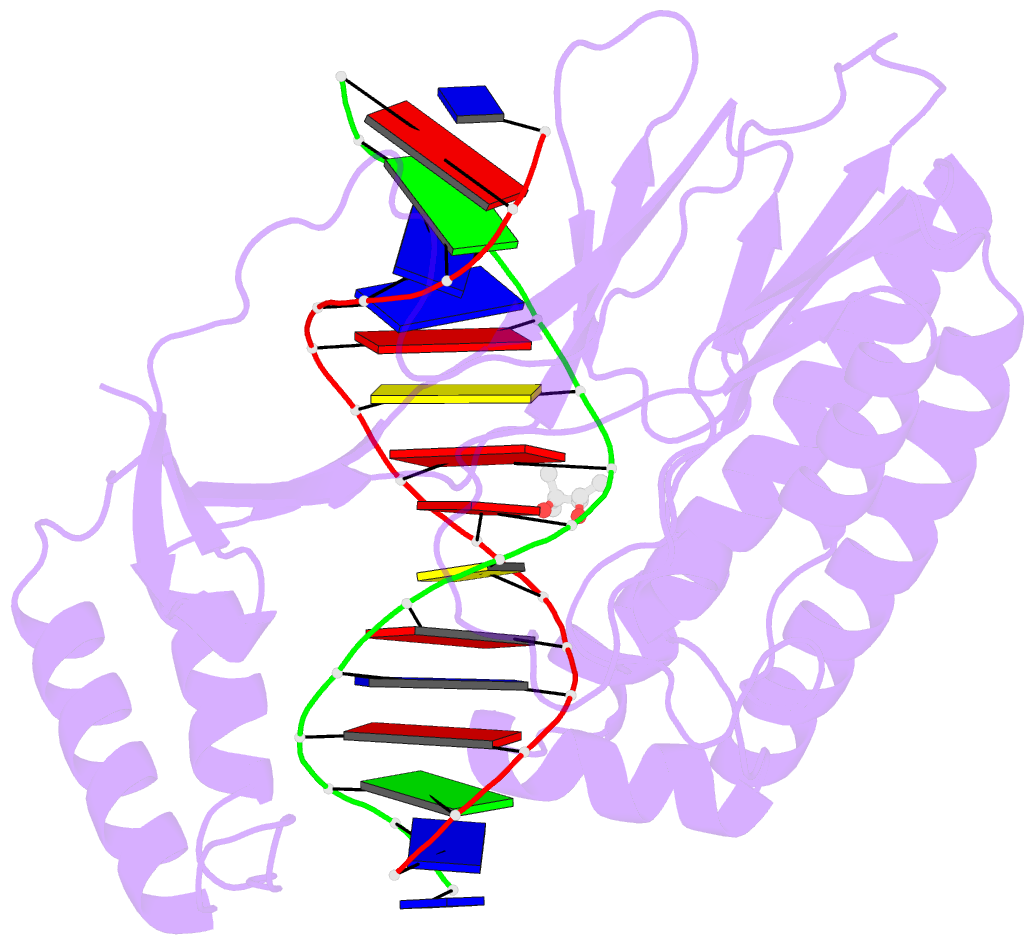
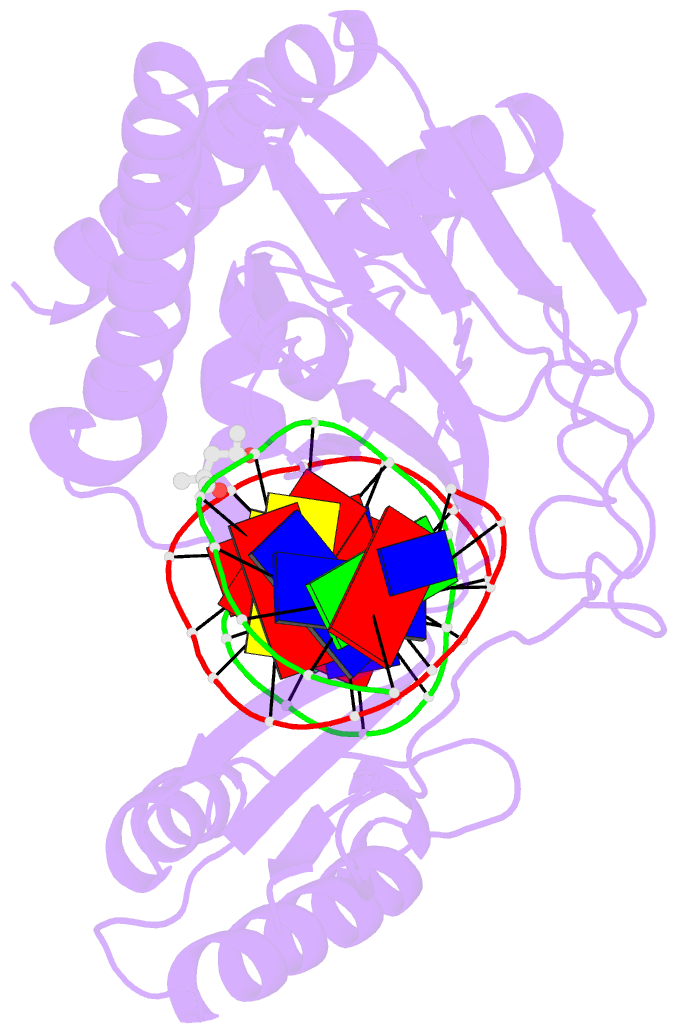
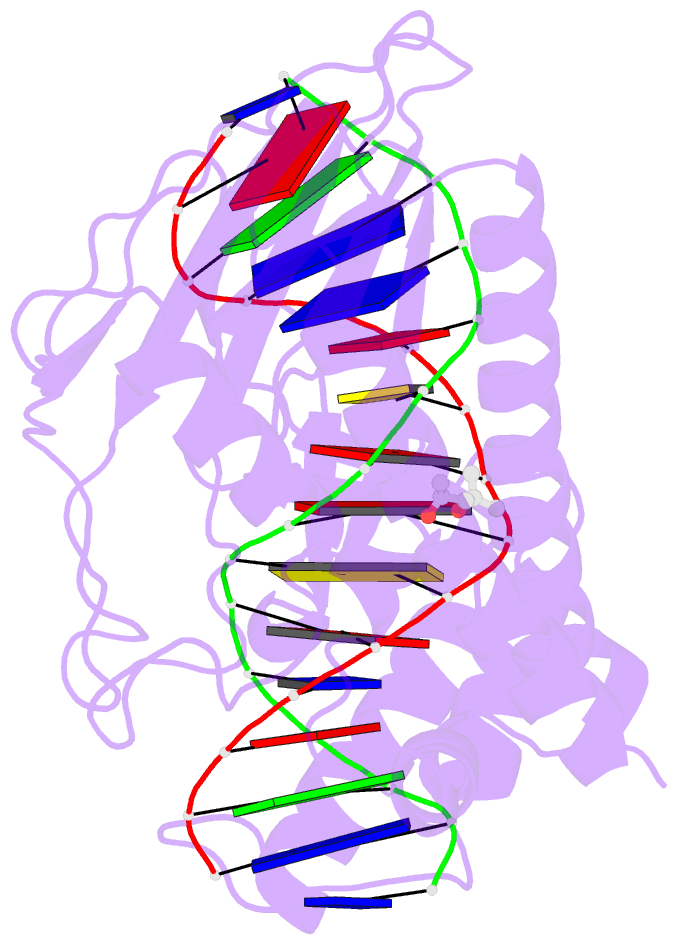
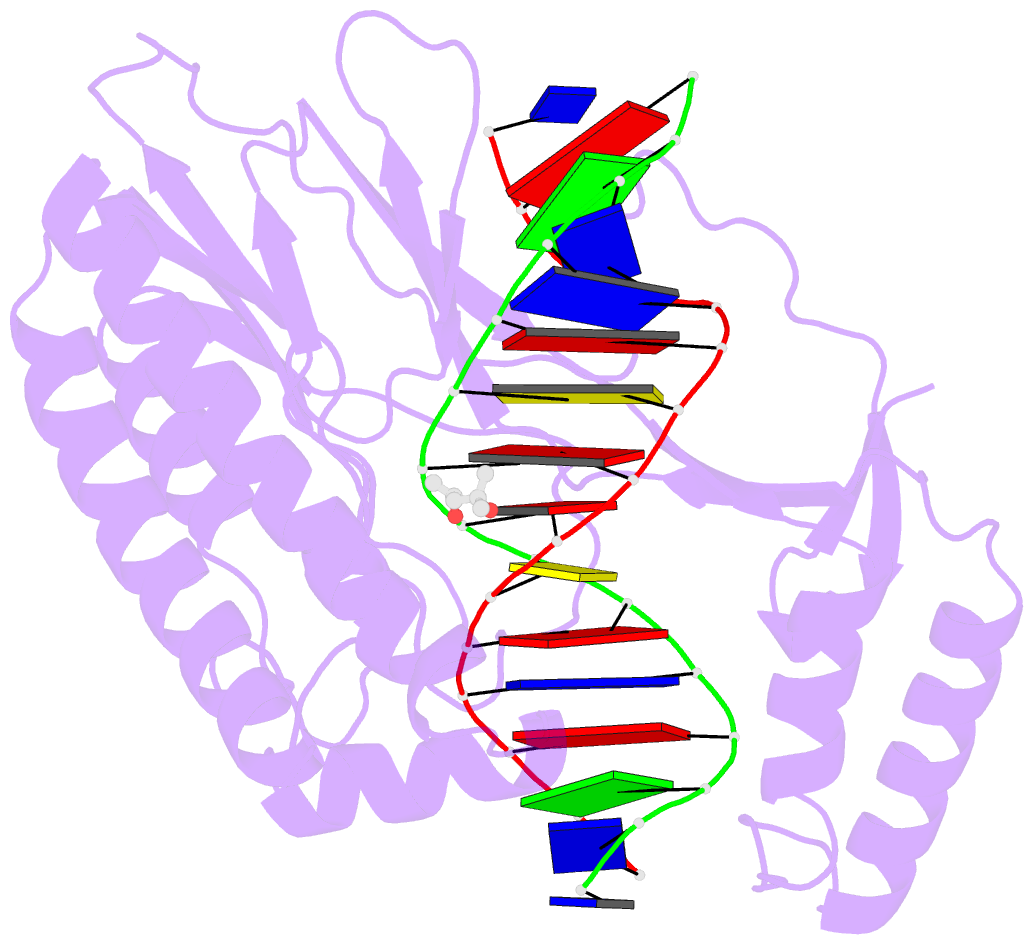
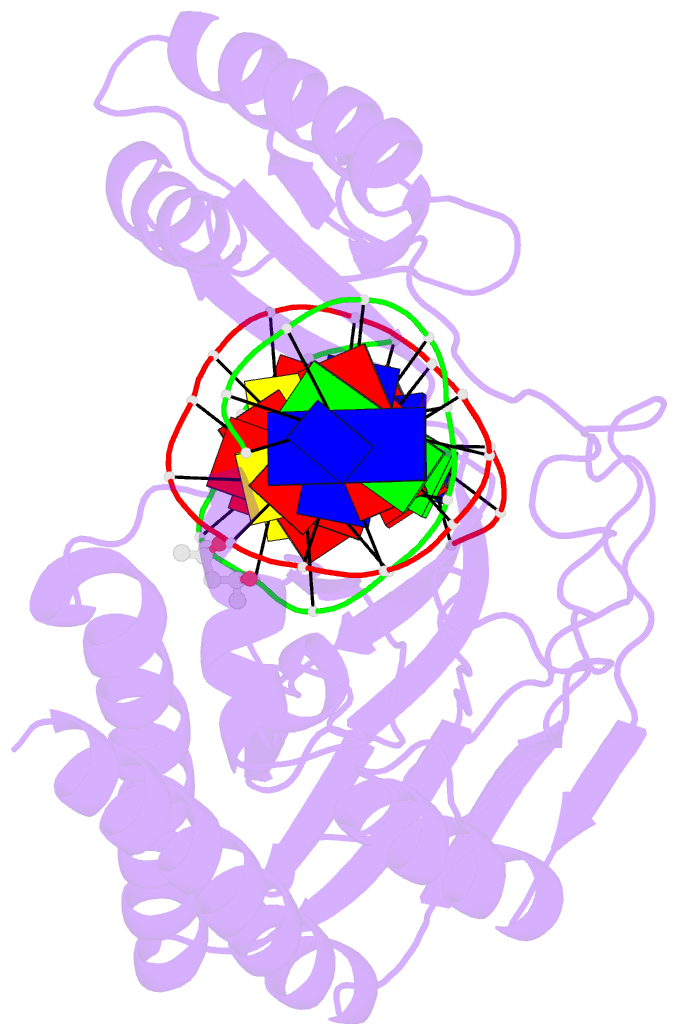
Summary information and primary citation
- PDB-id
- 4xr3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- replication-DNA
- Method
- X-ray (2.7 Å)
- Summary
- Escherichia coli replication terminator protein (tus) complexed with DNA- gc(6) swapped.
- Reference
- Elshenawy MM, Jergic S, Xu ZQ, Sobhy MA, Takahashi M, Oakley AJ, Dixon NE, Hamdan SM (2015): "Replisome speed determines the efficiency of the Tus-Ter replication termination barrier." Nature, 525, 394-398. doi: 10.1038/nature14866.
- Abstract
- In all domains of life, DNA synthesis occurs bidirectionally from replication origins. Despite variable rates of replication fork progression, fork convergence often occurs at specific sites. Escherichia coli sets a 'replication fork trap' that allows the first arriving fork to enter but not to leave the terminus region. The trap is set by oppositely oriented Tus-bound Ter sites that block forks on approach from only one direction. However, the efficiency of fork blockage by Tus-Ter does not exceed 50% in vivo despite its apparent ability to almost permanently arrest replication forks in vitro. Here we use data from single-molecule DNA replication assays and structural studies to show that both polarity and fork-arrest efficiency are determined by a competition between rates of Tus displacement and rearrangement of Tus-Ter interactions that leads to blockage of slower moving replisomes by two distinct mechanisms. To our knowledge this is the first example where intrinsic differences in rates of individual replisomes have different biological outcomes.