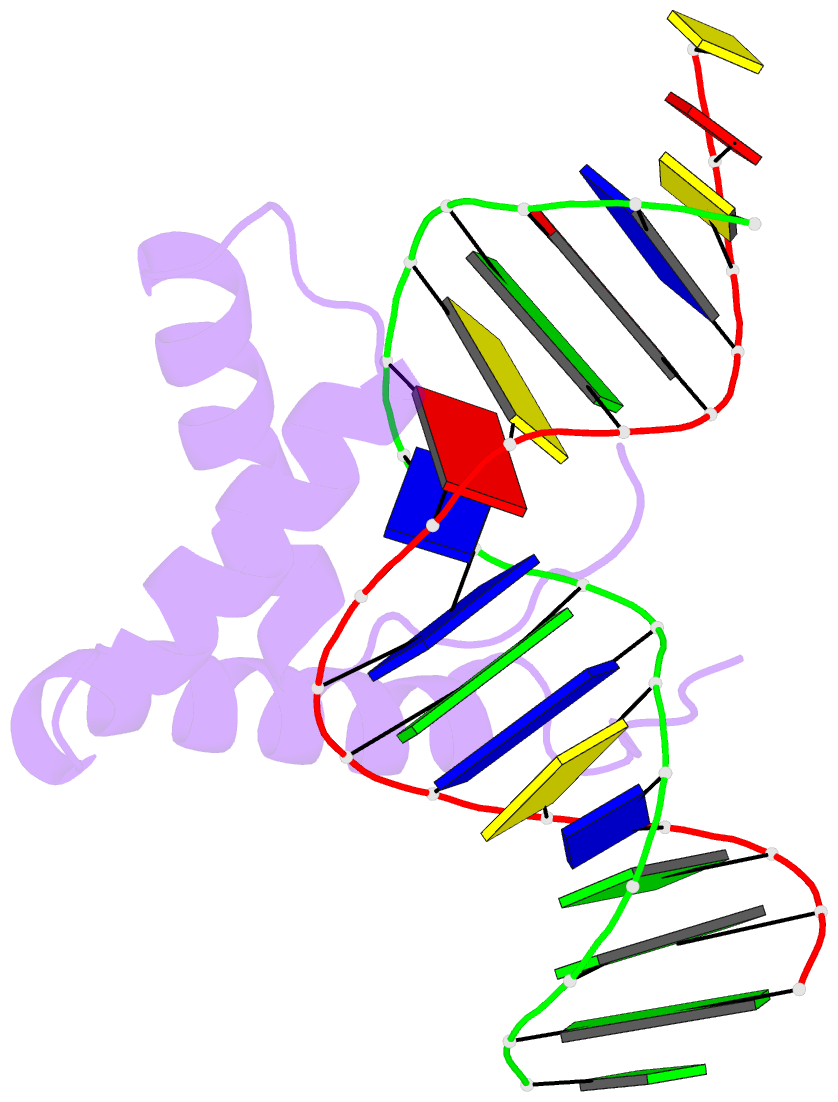
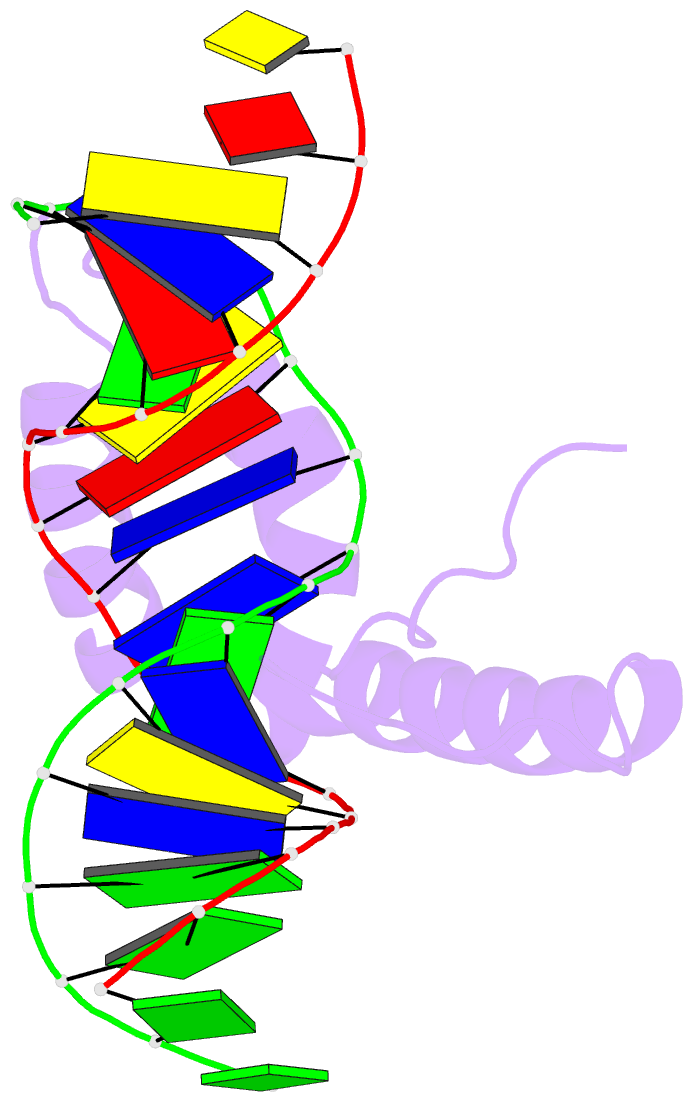
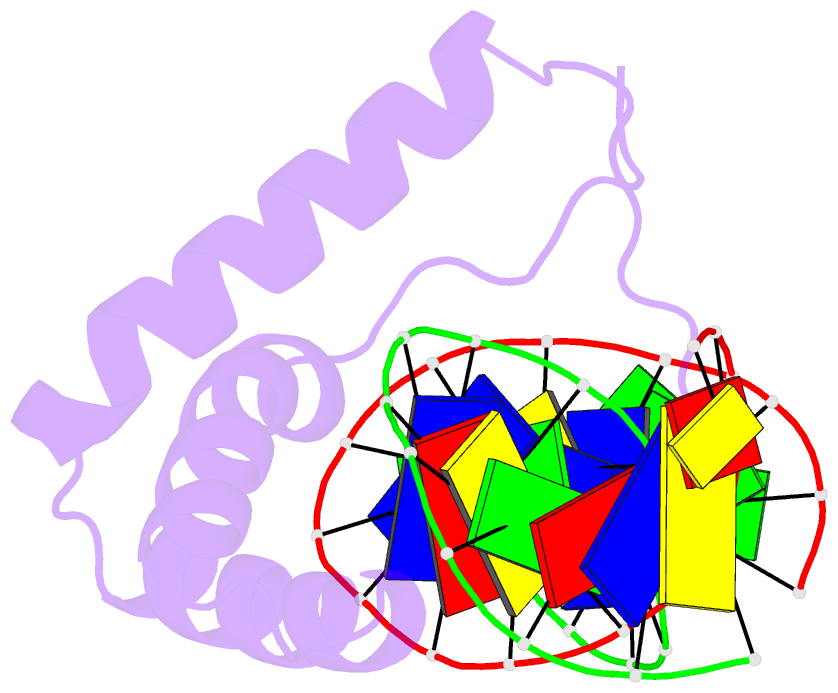
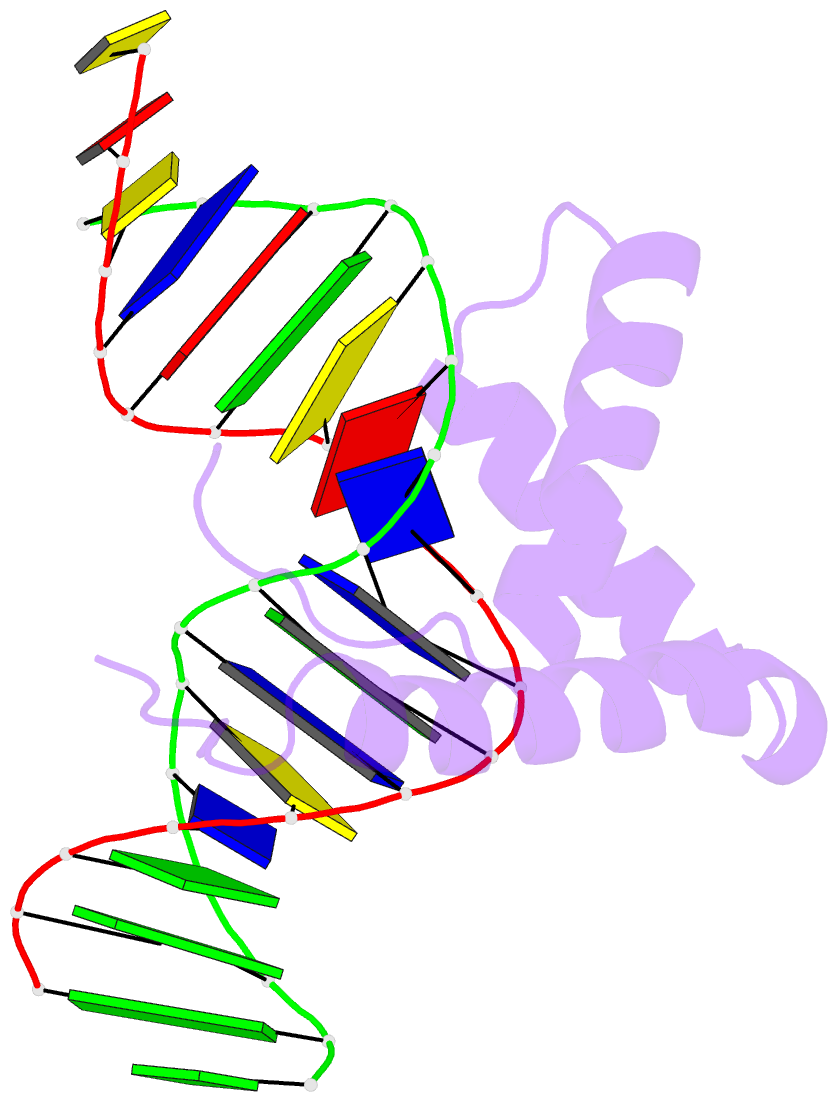
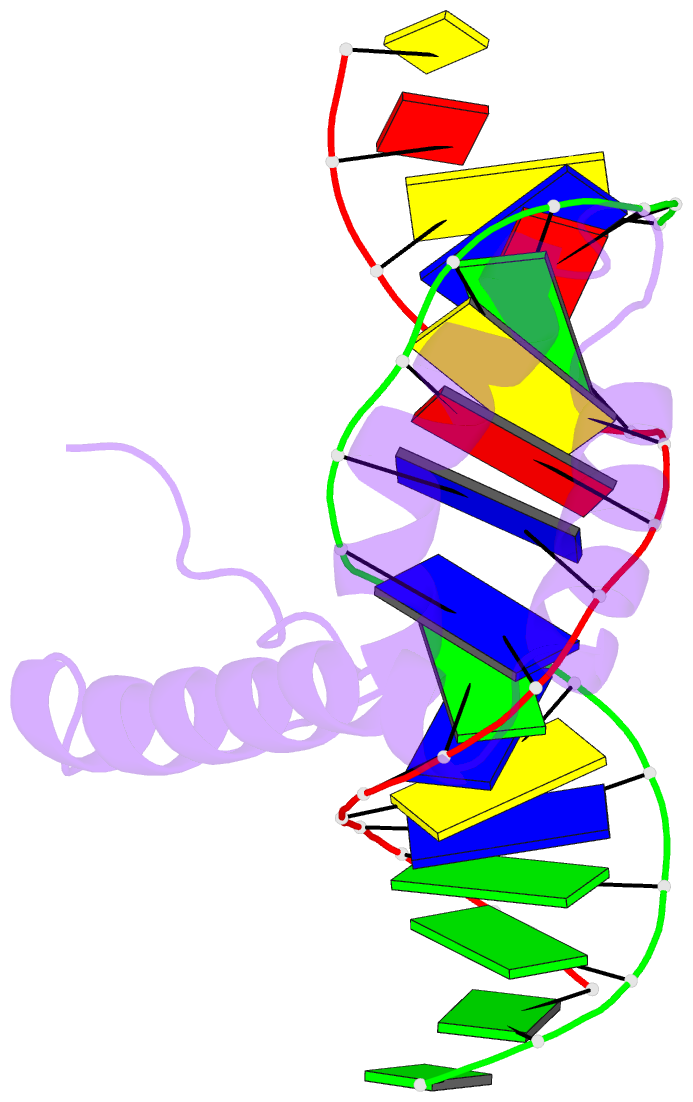
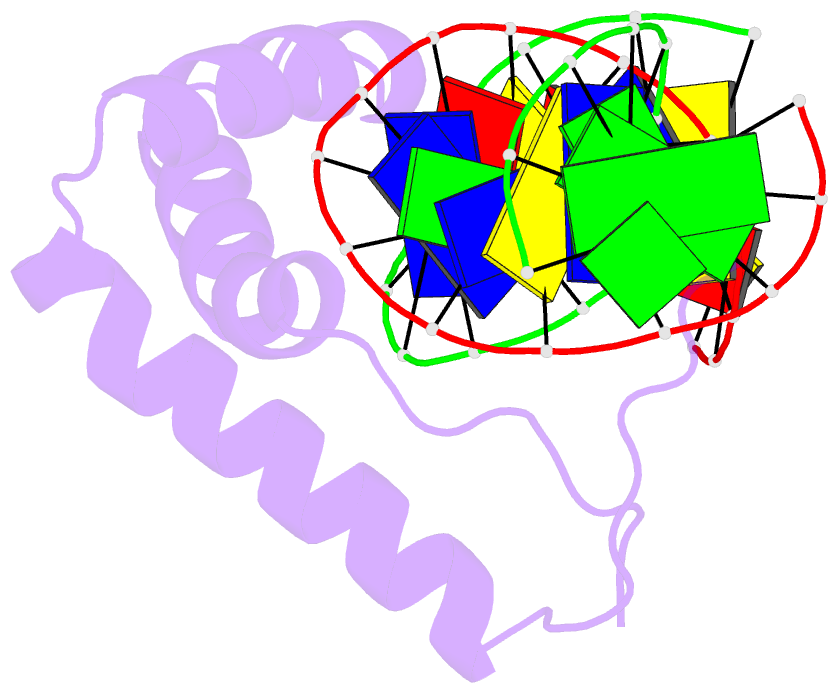
Summary information and primary citation
- PDB-id
- 4y60; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (1.75 Å)
- Summary
- Structure of sox18-hmg-prox1-DNA
- Reference
- Klaus M, Prokoph N, Girbig M, Wang X, Huang YH, Srivastava Y, Hou L, Narasimhan K, Kolatkar PR, Francois M, Jauch R (2016): "Structure and decoy-mediated inhibition of the SOX18/Prox1-DNA interaction." Nucleic Acids Res., 44, 3922-3935. doi: 10.1093/nar/gkw130.
- Abstract
- The transcription factor (TF) SOX18 drives lymphatic vessel development in both embryogenesis and tumour-induced neo-lymphangiogenesis. Genetic disruption of Sox18 in a mouse model protects from tumour metastasis and established the SOX18 protein as a molecular target. Here, we report the crystal structure of the SOX18 DNA binding high-mobility group (HMG) box bound to a DNA element regulating Prox1 transcription. The crystals diffracted to 1.75Å presenting the highest resolution structure of a SOX/DNA complex presently available revealing water structure, structural adjustments at the DNA contact interface and non-canonical conformations of the DNA backbone. To explore alternatives to challenging small molecule approaches for targeting the DNA-binding activity of SOX18, we designed a set of five decoys based on modified Prox1-DNA. Four decoys potently inhibited DNA binding of SOX18 in vitro and did not interact with non-SOX TFs. Serum stability, nuclease resistance and thermal denaturation assays demonstrated that a decoy circularized with a hexaethylene glycol linker and terminal phosphorothioate modifications is most stable. This SOX decoy also interfered with the expression of a luciferase reporter under control of a SOX18-dependent VCAM1 promoter in COS7 cells. Collectively, we propose SOX decoys as potential strategy for inhibiting SOX18 activity to disrupt tumour-induced neo-lymphangiogenesis.