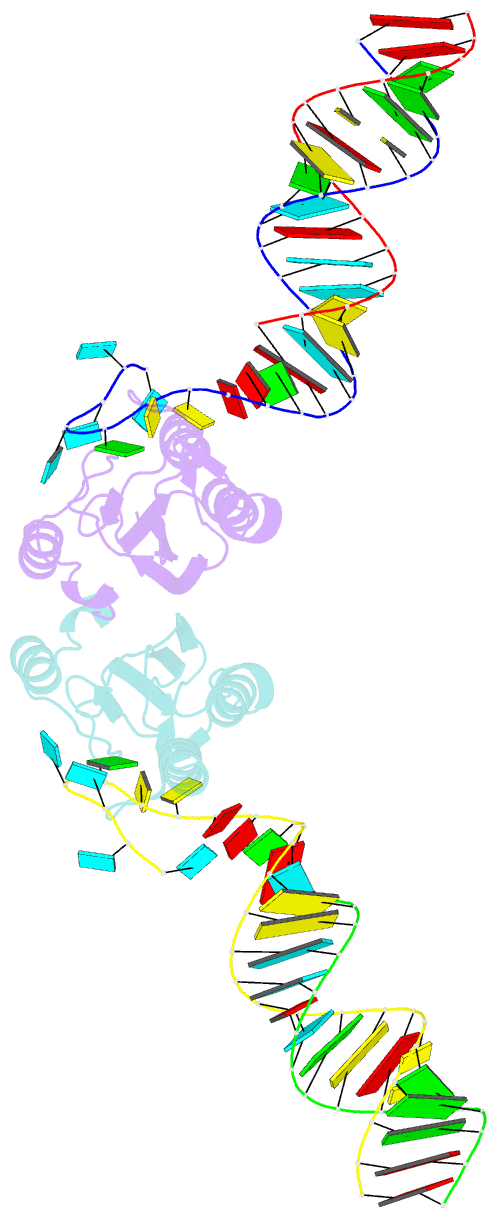
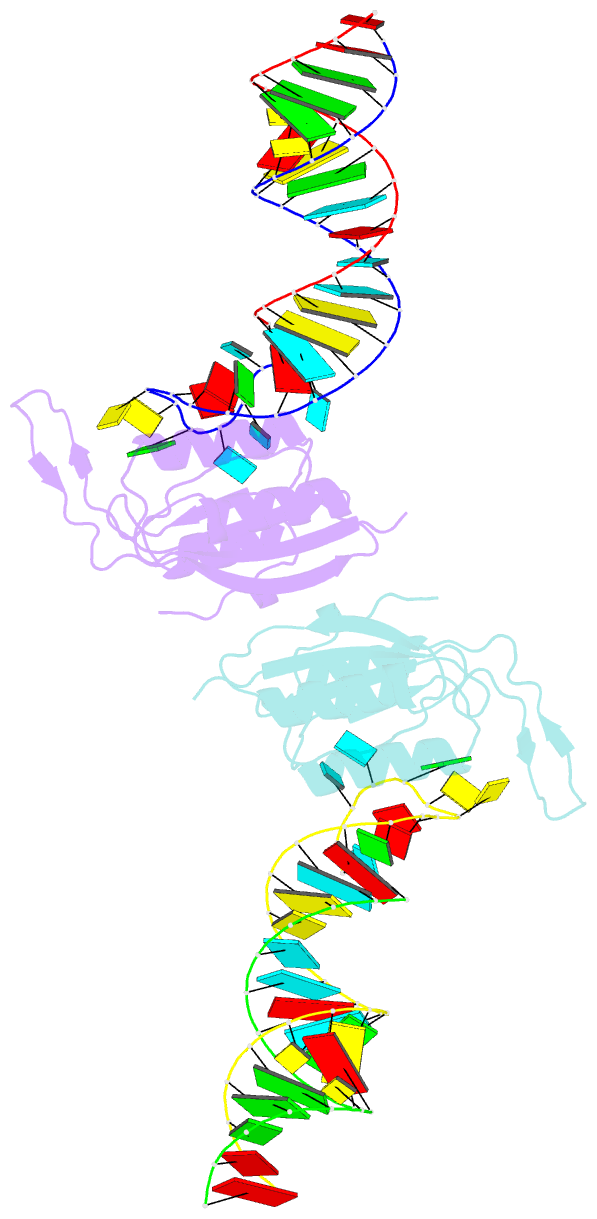
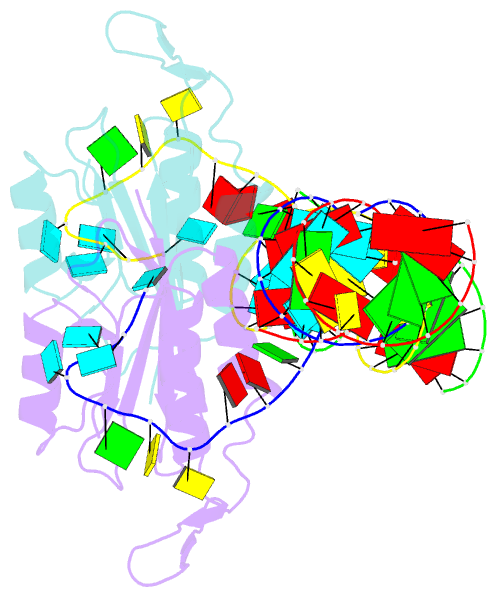
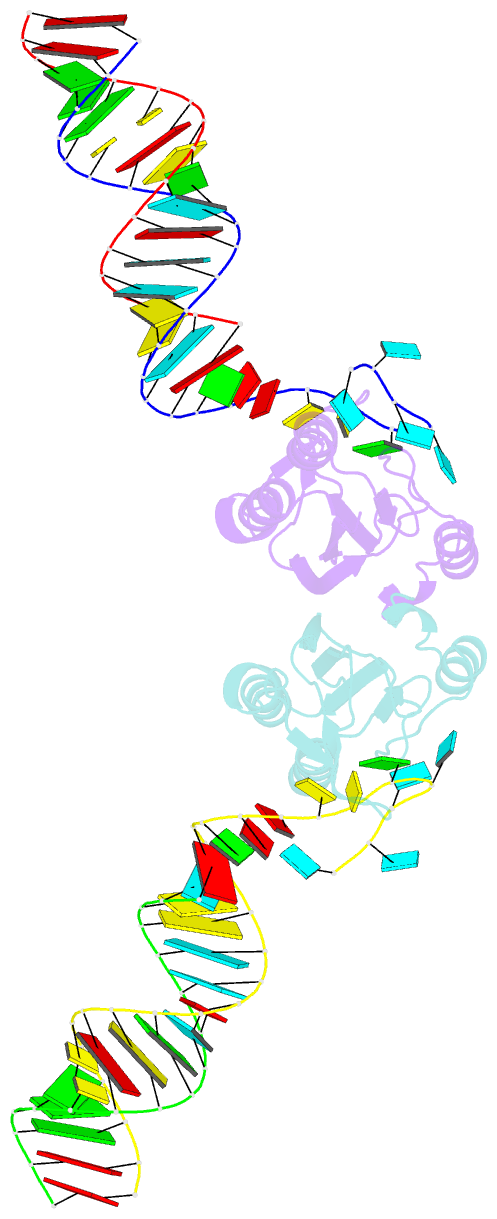
Summary information and primary citation
- PDB-id
- 4yhw; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- splicing
- Method
- X-ray (3.25 Å)
- Summary
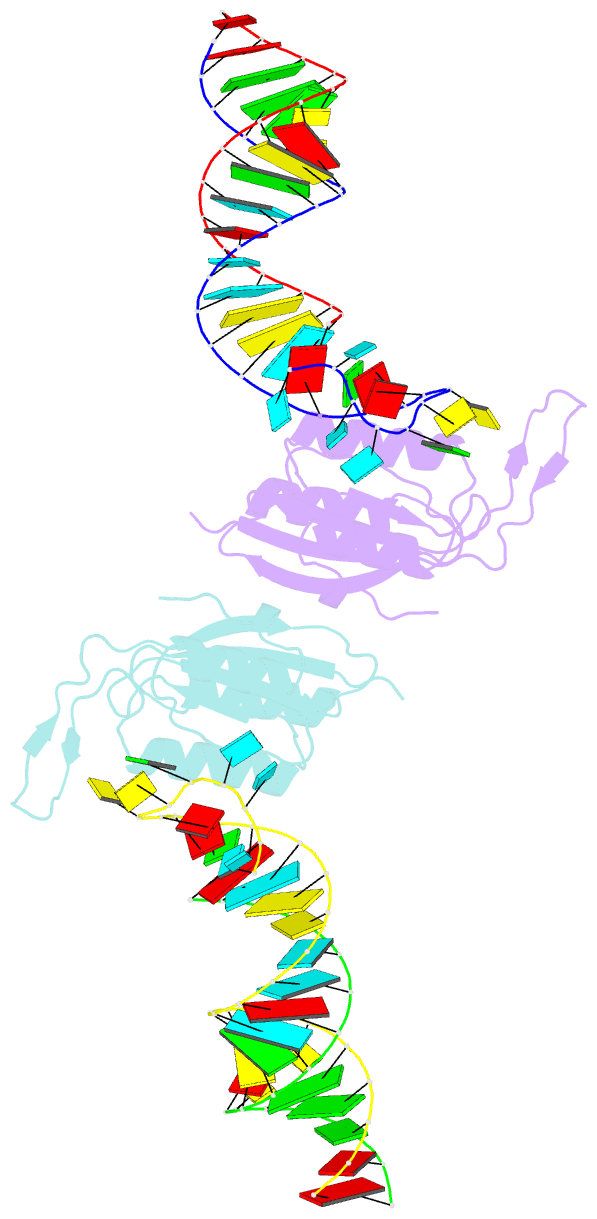
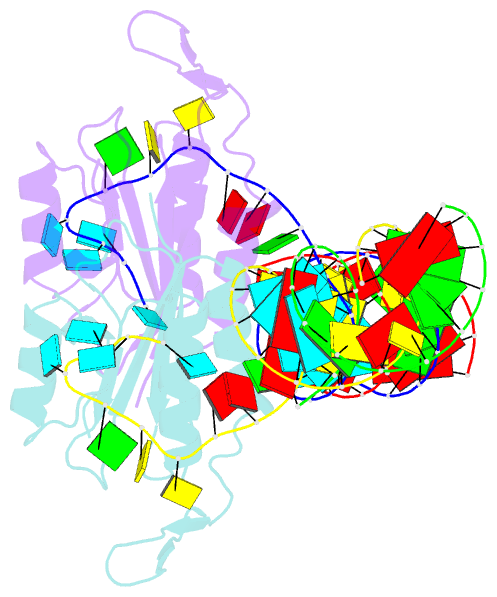
- Yeast prp3 (296-469) in complex with fragment of u4-u6 di-snrna
- Reference
- Liu S, Mozaffari-Jovin S, Wollenhaupt J, Santos KF, Theuser M, Dunin-Horkawicz S, Fabrizio P, Bujnicki JM, Luhrmann R, Wahl MC (2015): "A composite double-/single-stranded RNA-binding region in protein Prp3 supports tri-snRNP stability and splicing." Elife, 4, e07320. doi: 10.7554/eLife.07320.
- Abstract
- Prp3 is an essential U4/U6 di-snRNP-associated protein whose functions and molecular mechanisms in pre-mRNA splicing are presently poorly understood. We show by structural and biochemical analyses that Prp3 contains a bipartite U4/U6 di-snRNA-binding region comprising an expanded ferredoxin-like fold, which recognizes a 3'-overhang of U6 snRNA, and a preceding peptide, which binds U4/U6 stem II. Phylogenetic analyses revealed that the single-stranded RNA-binding domain is exclusively found in Prp3 orthologs, thus qualifying as a spliceosome-specific RNA interaction module. The composite double-stranded/single-stranded RNA-binding region assembles cooperatively with Snu13 and Prp31 on U4/U6 di-snRNAs and inhibits Brr2-mediated U4/U6 di-snRNA unwinding in vitro. RNP-disrupting mutations in Prp3 lead to U4/U6•U5 tri-snRNP assembly and splicing defects in vivo. Our results reveal how Prp3 acts as an important bridge between U4/U6 and U5 in the tri-snRNP and comparison with a Prp24-U6 snRNA recycling complex suggests how Prp3 may be involved in U4/U6 reassembly after splicing.