Summary information and primary citation
- PDB-id
- 4yow; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.5 Å)
- Summary
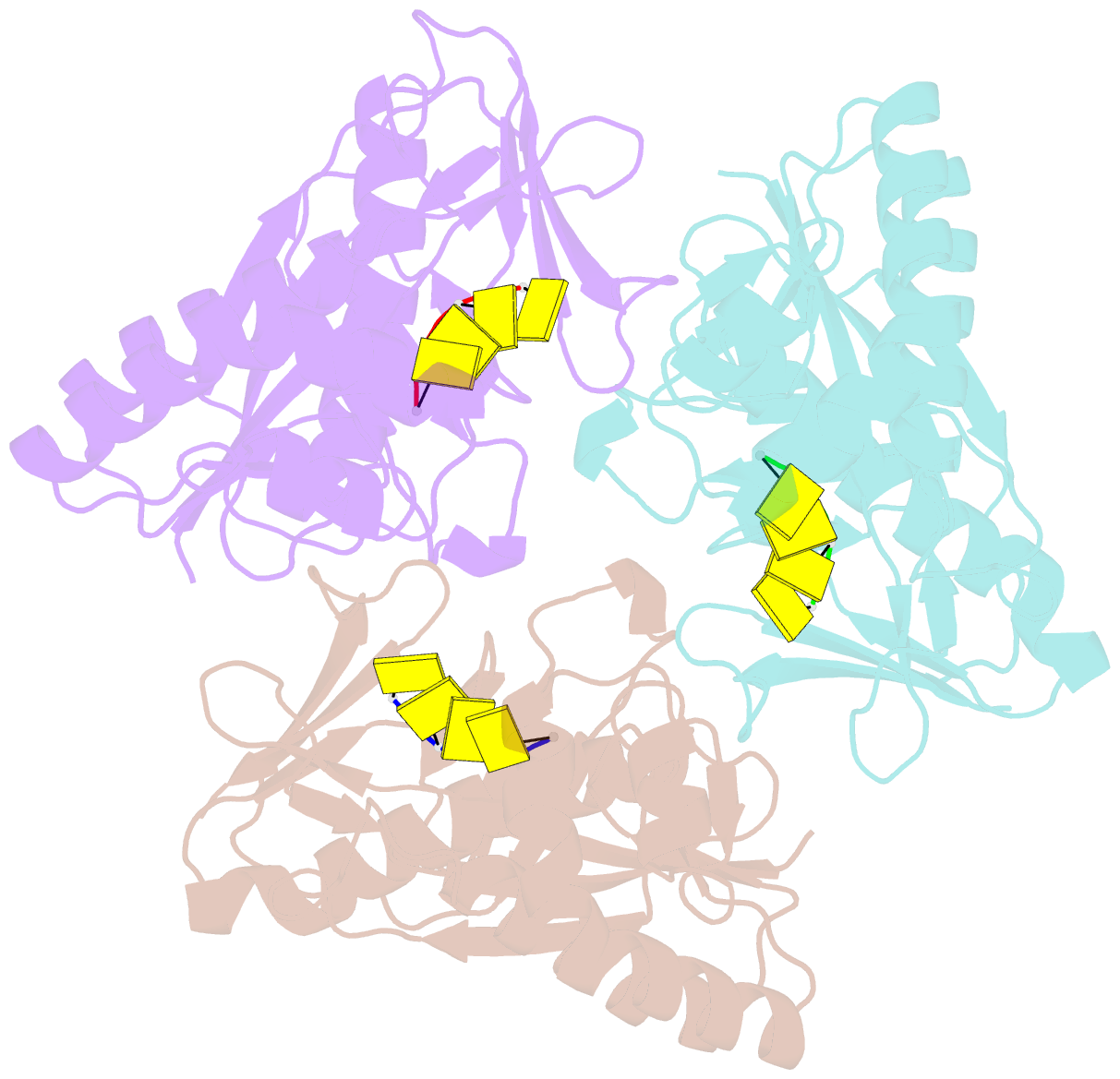
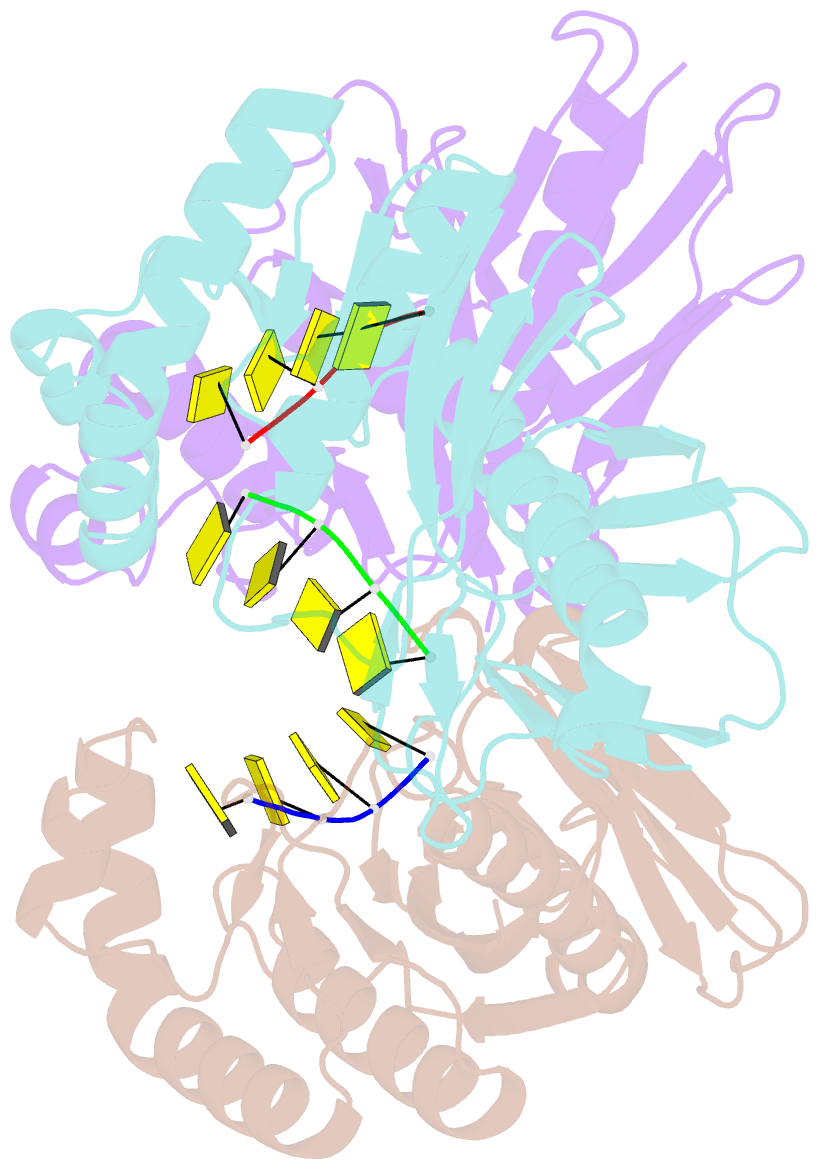
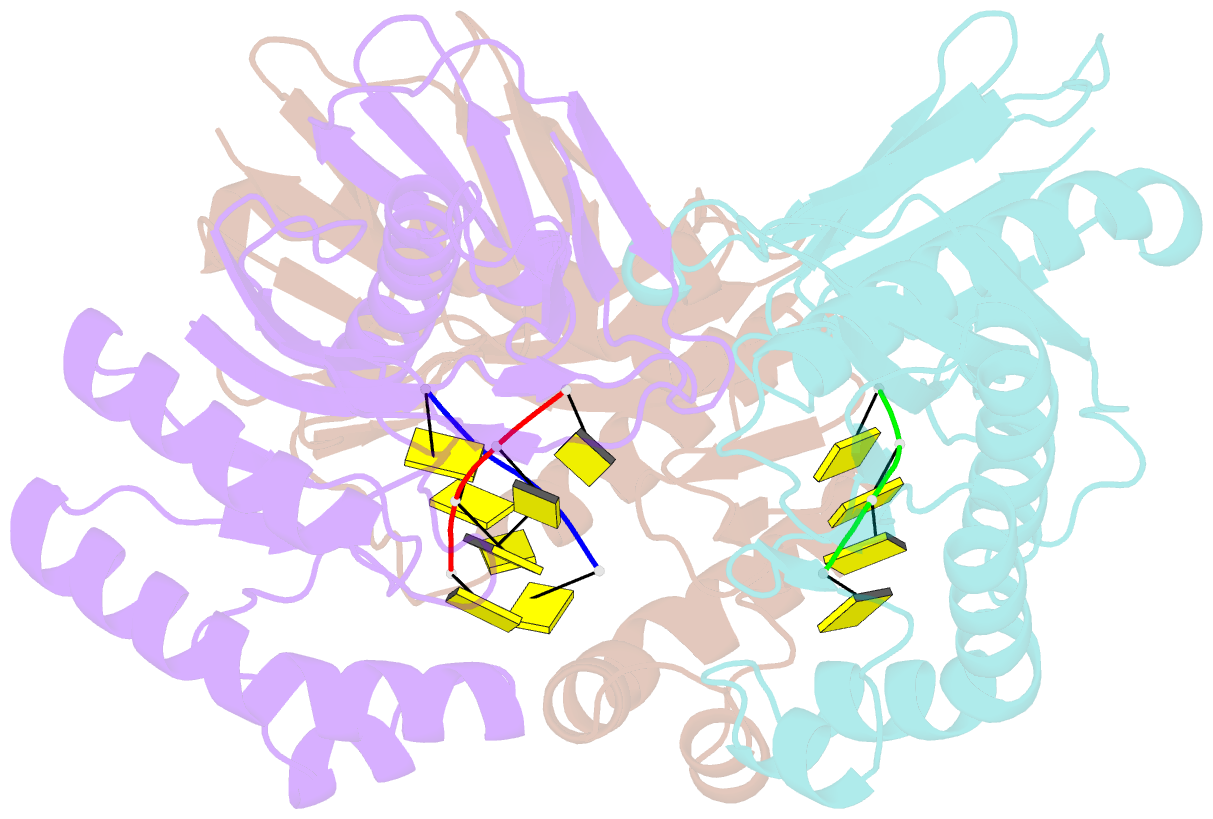
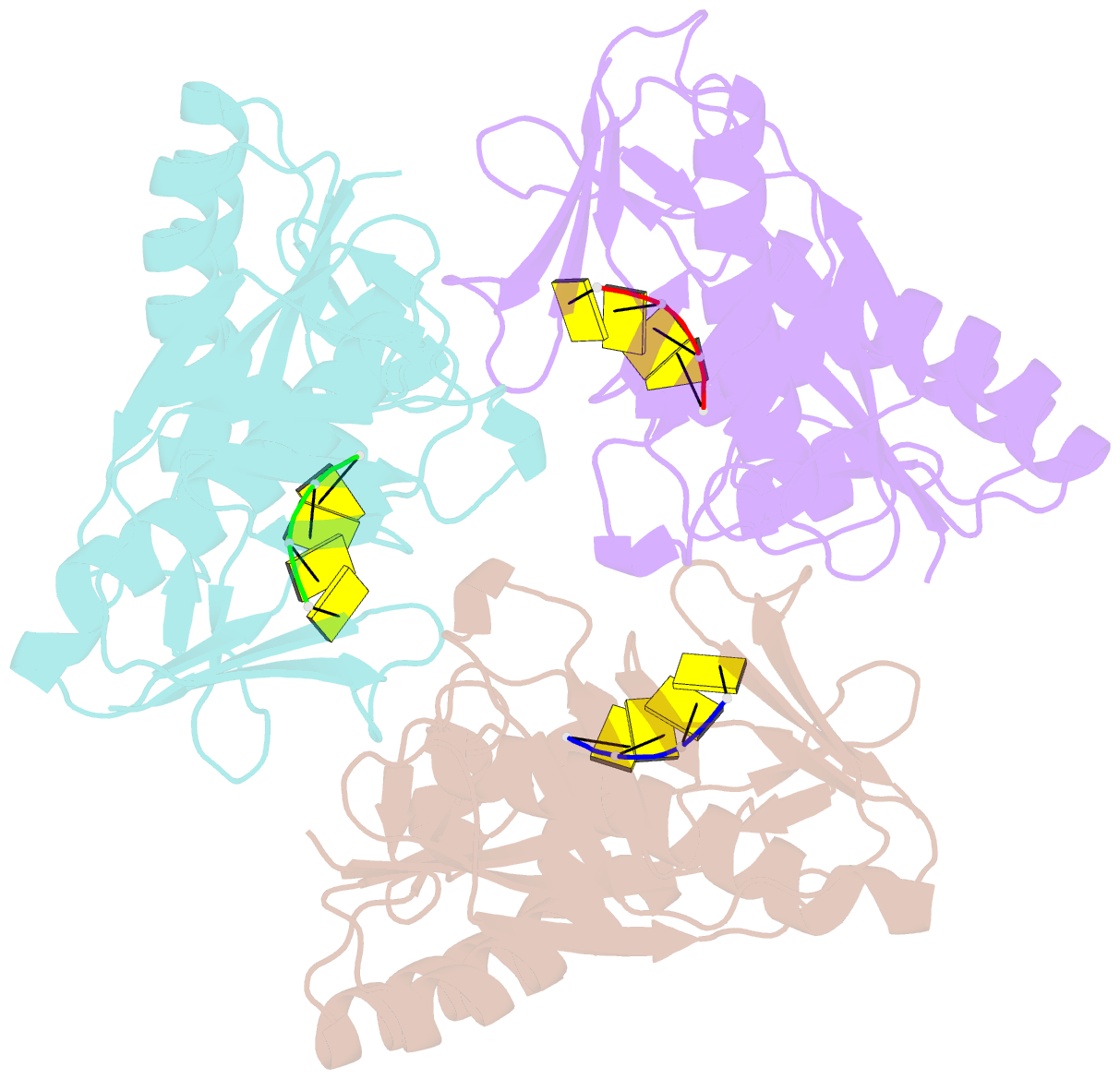
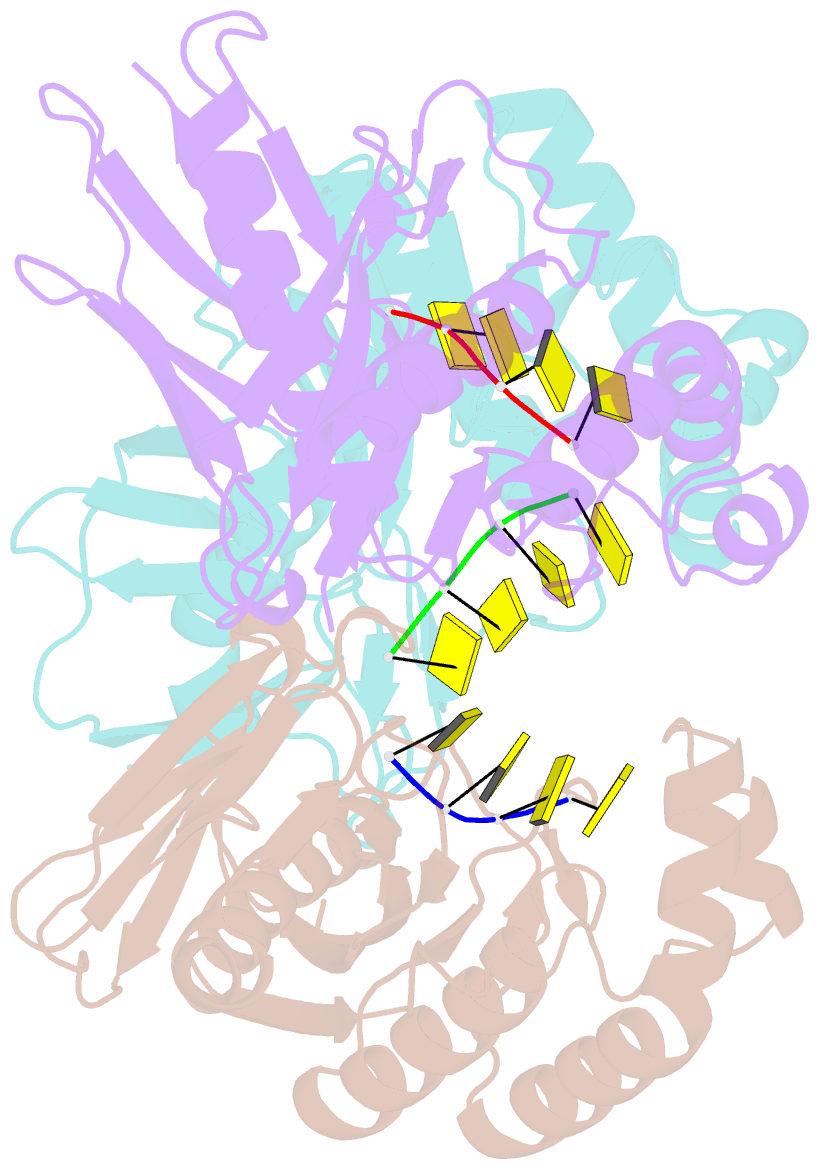
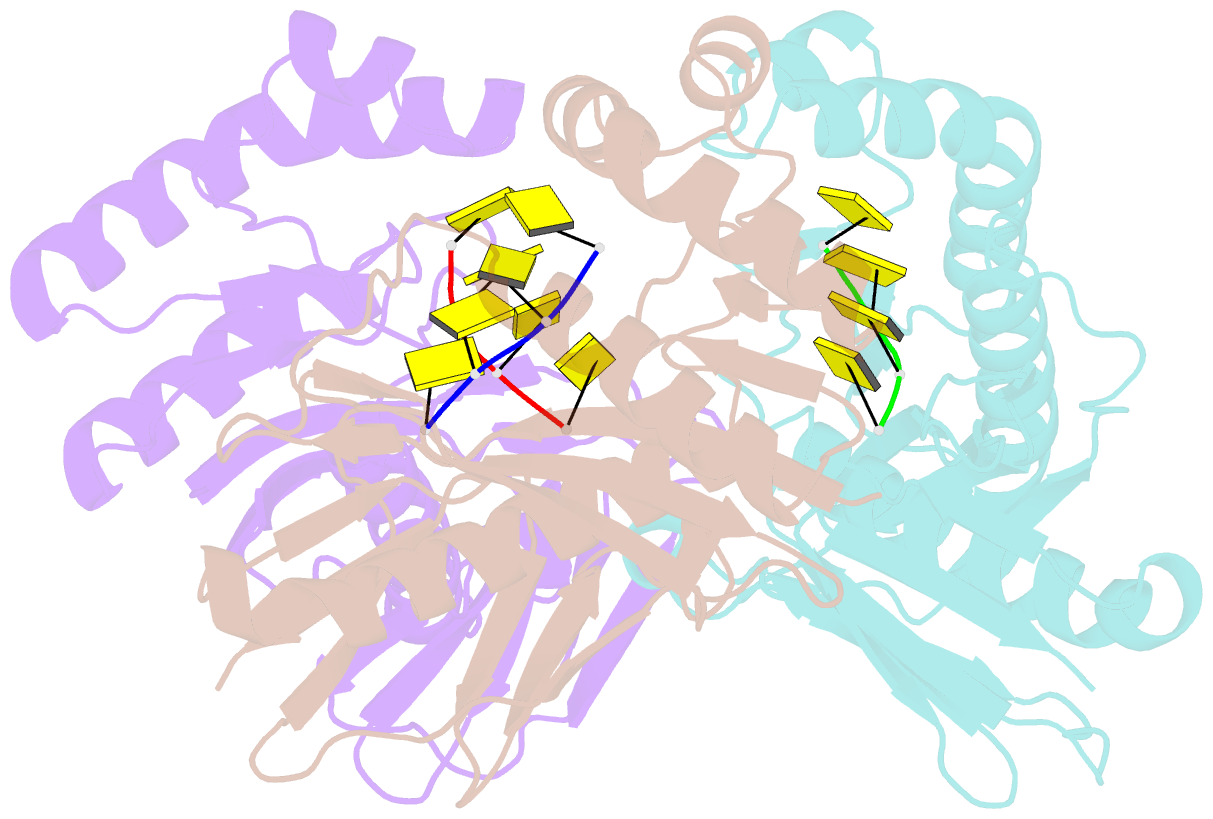
- Crystal structure of a trimeric exonuclease phoexo i from pyrococcus horikoshii ot3 in complex with poly-dc
- Reference
- Miyazono K, Ishino S, Tsutsumi K, Ito T, Ishino Y, Tanokura M (2015): "Structural basis for substrate recognition and processive cleavage mechanisms of the trimeric exonuclease PhoExo I." Nucleic Acids Res., 43, 7122-7136. doi: 10.1093/nar/gkv654.
- Abstract
- Nucleases play important roles in nucleic acid processes, such as replication, repair and recombination. Recently, we identified a novel single-strand specific 3'-5' exonuclease, PfuExo I, from the hyperthermophilic archaeon Pyrococcus furiosus, which may be involved in the Thermococcales-specific DNA repair system. PfuExo I forms a trimer and cleaves single-stranded DNA at every two nucleotides. Here, we report the structural basis for the cleavage mechanism of this novel exonuclease family. A structural analysis of PhoExo I, the homologous enzyme from P. horikoshii OT3, showed that PhoExo I utilizes an RNase H-like active site and possesses a 3'-OH recognition site ∼9 Å away from the active site, which enables cleavage at every two nucleotides. Analyses of the heterotrimeric and monomeric PhoExo I activities showed that trimerization is indispensable for its processive cleavage mechanism, but only one active site of the trimer is required.