Summary information and primary citation
- PDB-id
- 4yye; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ligase-RNA
- Method
- X-ray (2.301 Å)
- Summary
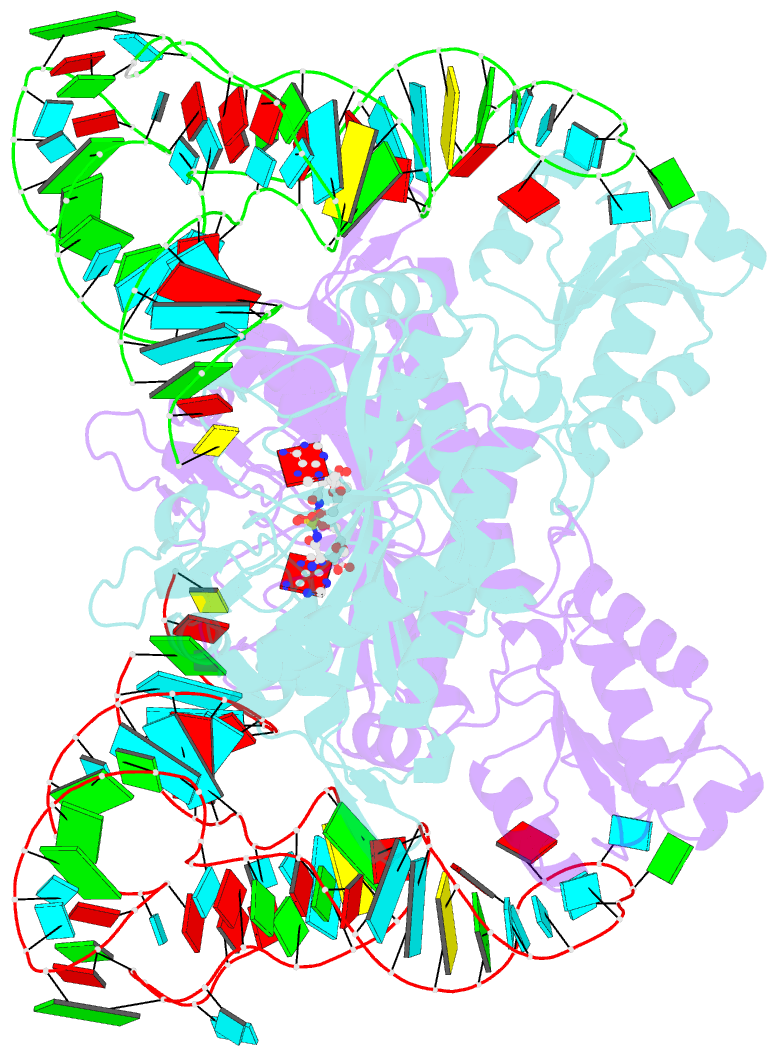
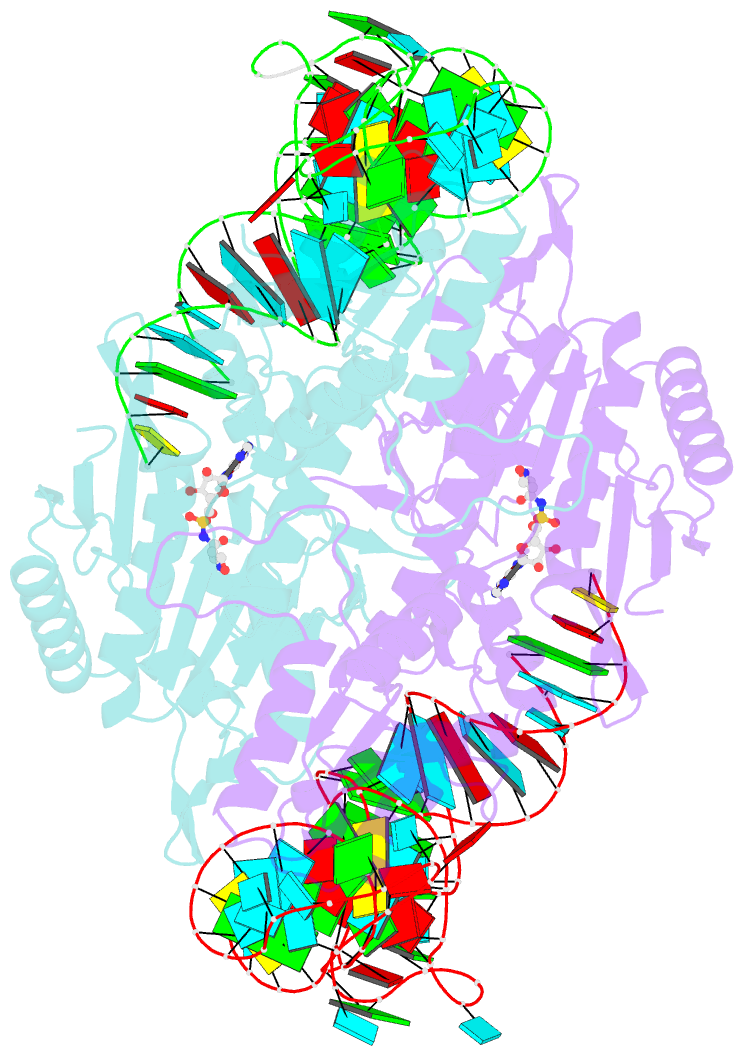
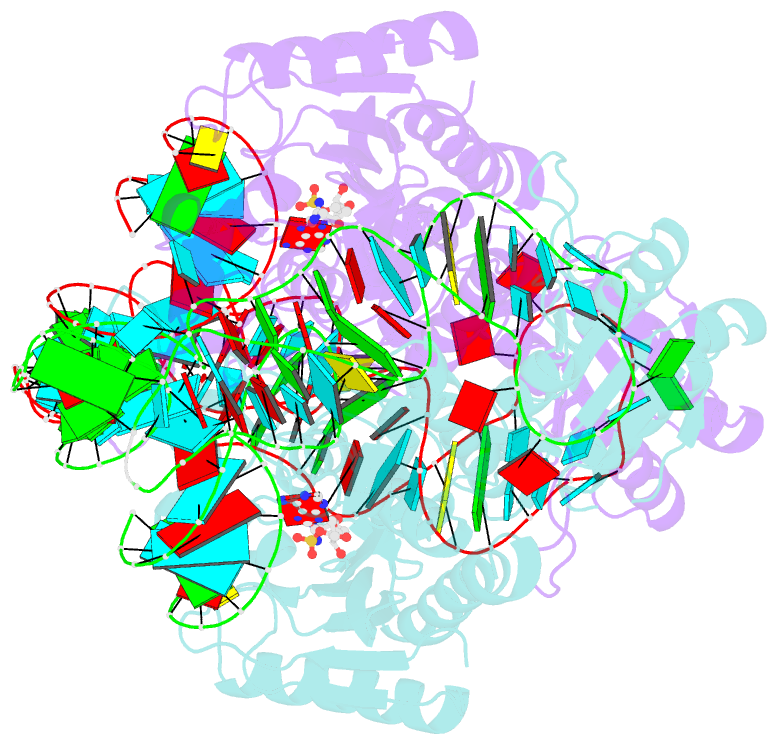
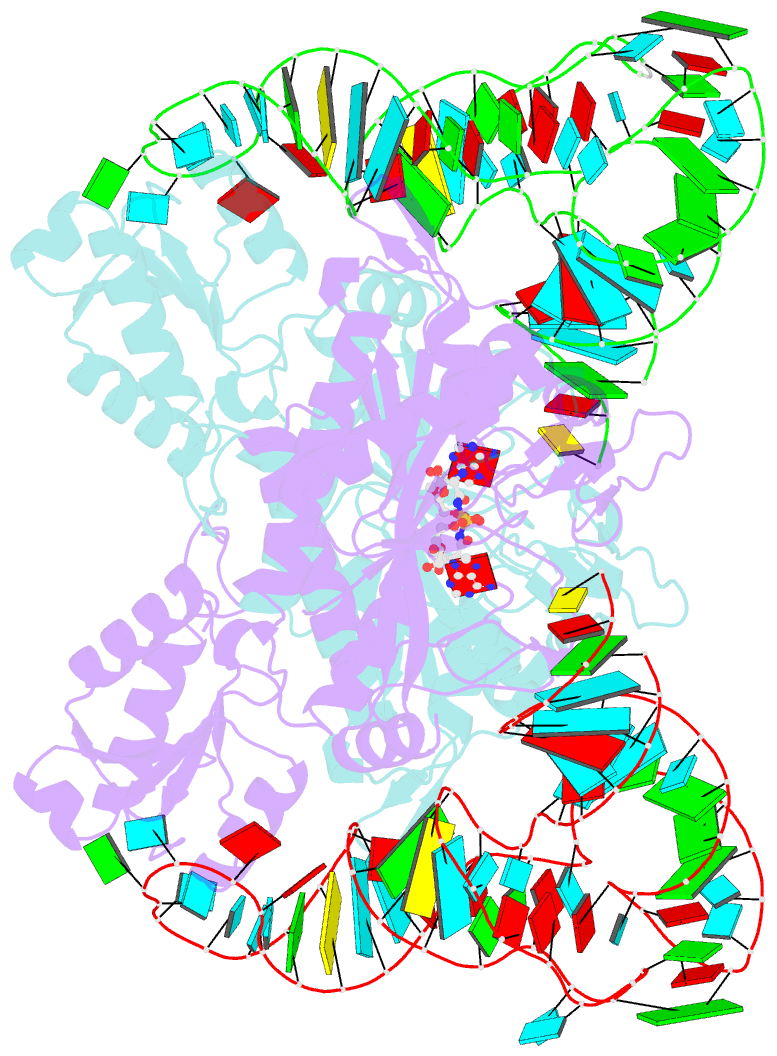
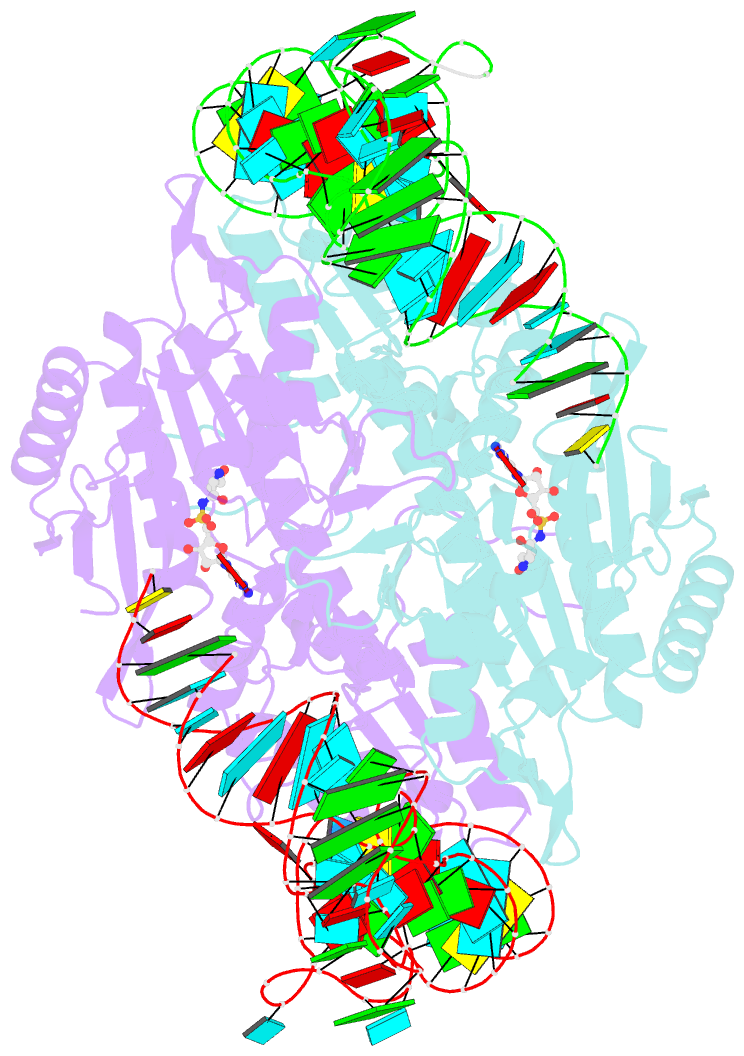
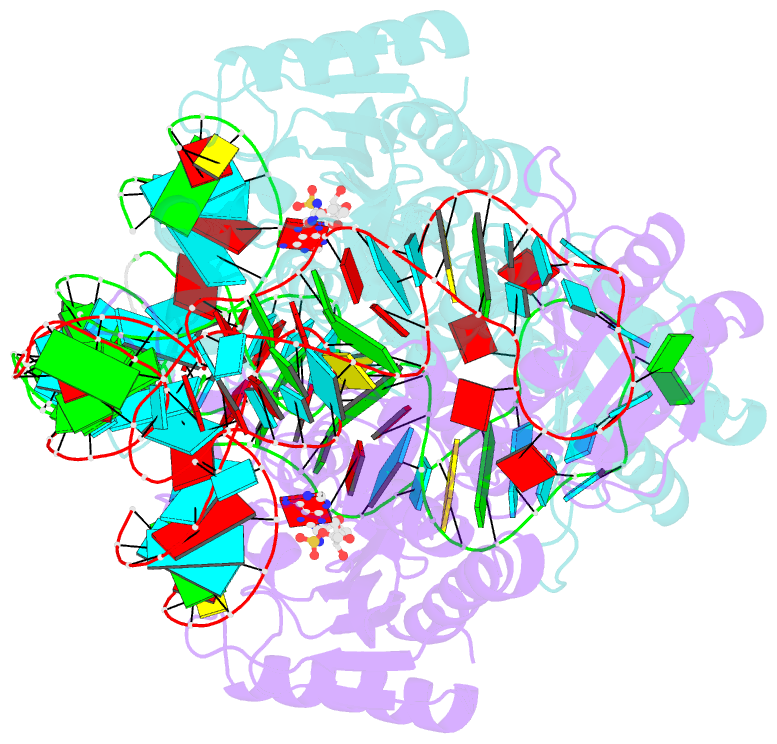
- Crystal structure of the yeast mitochondrial threonyl-trna synthetase (mst1) in complex with the canonical trnathr and threonyl sulfamoyl adenylate
- Reference
- Holman KM, Wu J, Ling J, Simonovic M (2016): "The crystal structure of yeast mitochondrial ThrRS in complex with the canonical threonine tRNA." Nucleic Acids Res., 44, 1428-1439. doi: 10.1093/nar/gkv1501.
- Abstract
- In mitochondria of Saccharomyces cerevisiae, a single aminoacyl-tRNA synthetase (aaRS), MST1, aminoacylates two isoacceptor tRNAs, tRNA1(Thr) and tRNA2(Thr), that harbor anticodon loops of different size and sequence. As a result of this promiscuity, reassignment of the CUN codon box from leucine to threonine is facilitated. However, the mechanism by which a single aaRS binds distinct anticodon loops with high specificity is not well understood. Herein, we present the crystal structure of MST1 in complex with the canonical tRNA2(Thr) and non-hydrolyzable analog of threonyl adenylate. Our structure reveals that the dimeric arrangement of MST1 is essential for binding the 5'-phosphate, the second base pair of the acceptor stem, the first two base pairs of the anticodon stem and the first nucleotide of the variable arm. Further, in contrast to the bacterial ortholog that 'reads' the entire anticodon sequence, MST1 recognizes bases in the second and third position and the nucleotide upstream of the anticodon sequence. We speculate that a flexible loop linking strands β4 and β5 may be allosteric regulator that establishes cross-subunit communication between the aminoacylation and tRNA-binding sites. We also propose that structural features of the anticodon-binding domain in MST1 permit binding of the enlarged anticodon loop of tRNA1(Thr).