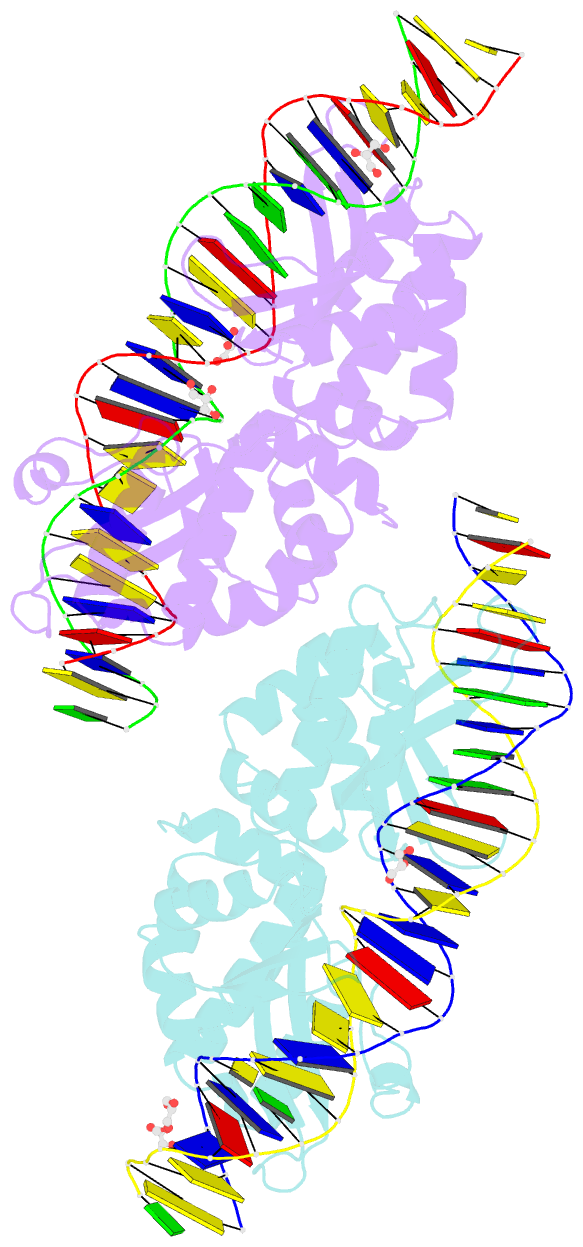
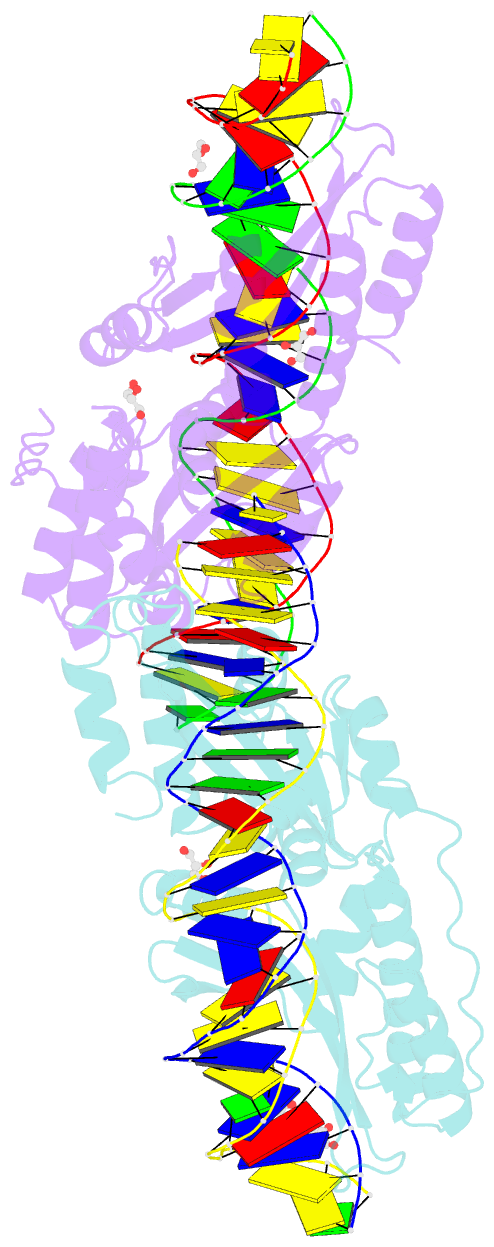
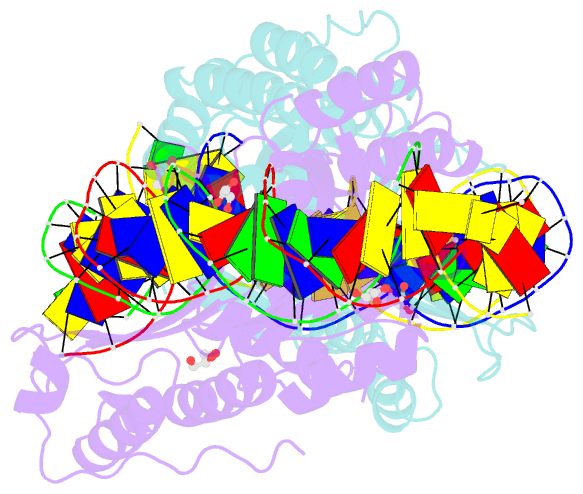
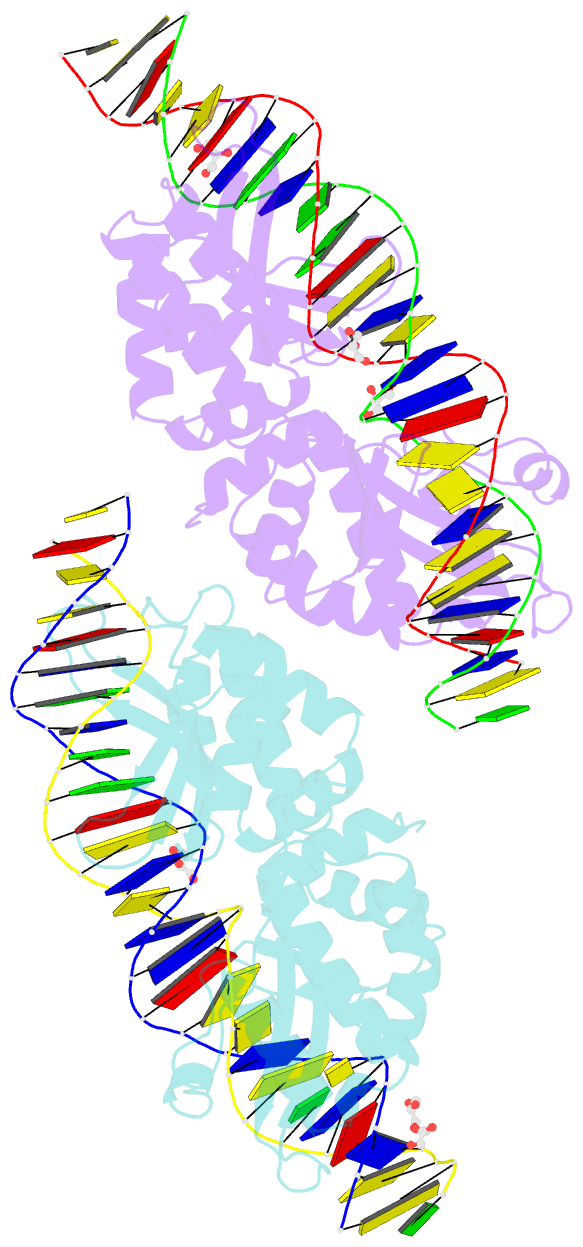
Summary information and primary citation
- PDB-id
- 4z1z; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (3.2 Å)
- Summary
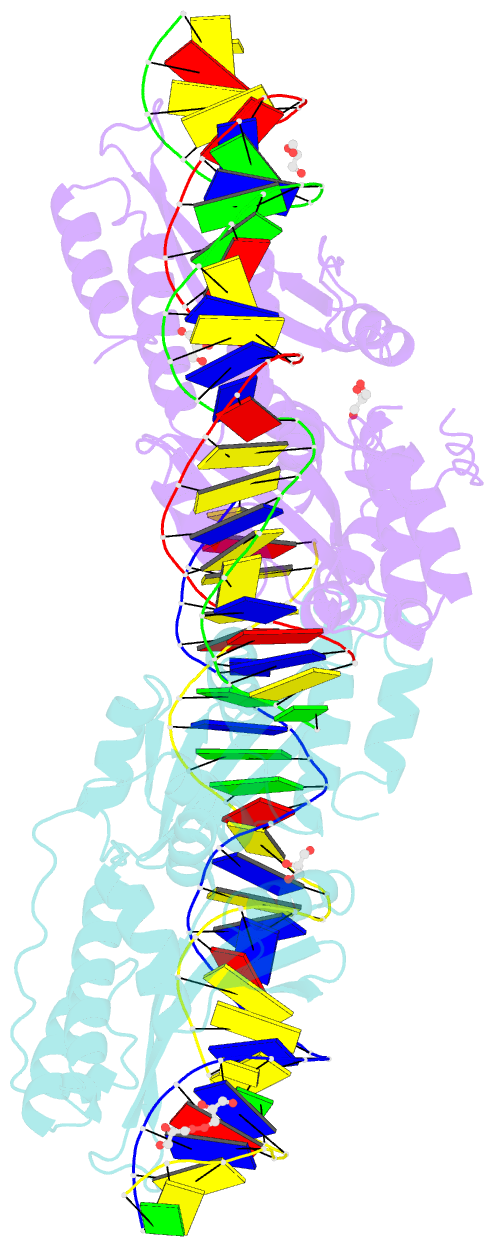
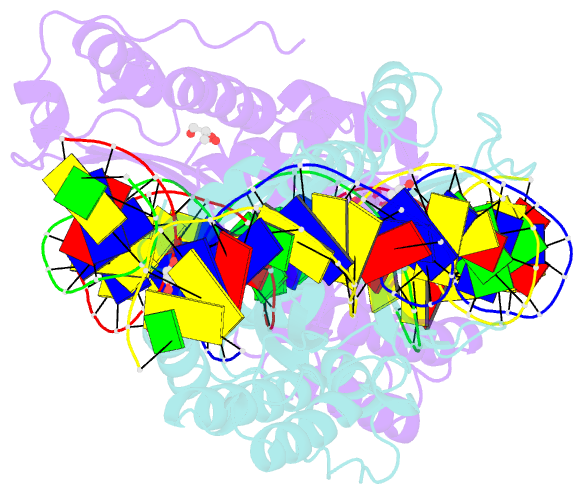
- Crystal structure of meganuclease i-smami bound to uncleaveable DNA with a ttct central four
- Reference
- Lambert AR, Hallinan JP, Shen BW, Chik JK, Bolduc JM, Kulshina N, Robins LI, Kaiser BK, Jarjour J, Havens K, Scharenberg AM, Stoddard BL (2016): "Indirect DNA Sequence Recognition and Its Impact on Nuclease Cleavage Activity." Structure, 24, 862-873. doi: 10.1016/j.str.2016.03.024.
- Abstract
- LAGLIDADG meganucleases are DNA cleaving enzymes used for genome engineering. While their cleavage specificity can be altered using several protein engineering and selection strategies, their overall targetability is limited by highly specific indirect recognition of the central four base pairs within their recognition sites. In order to examine the physical basis of indirect sequence recognition and to expand the number of such nucleases available for genome engineering, we have determined the target sites, DNA-bound structures, and central four cleavage fidelities of nine related enzymes. Subsequent crystallographic analyses of a meganuclease bound to two noncleavable target sites, each containing a single inactivating base pair substitution at its center, indicates that a localized slip of the mutated base pair causes a small change in the DNA backbone conformation that results in a loss of metal occupancy at one binding site, eliminating cleavage activity.