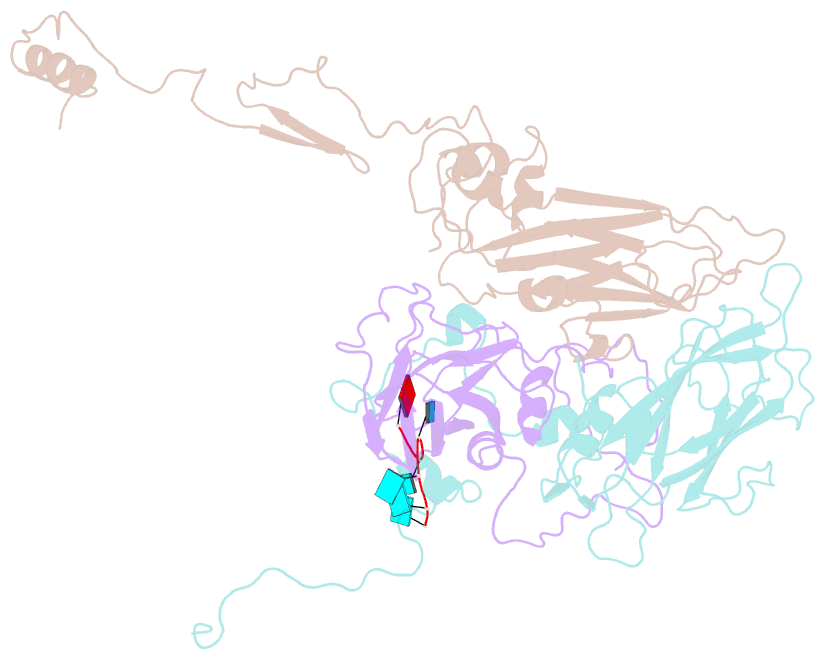
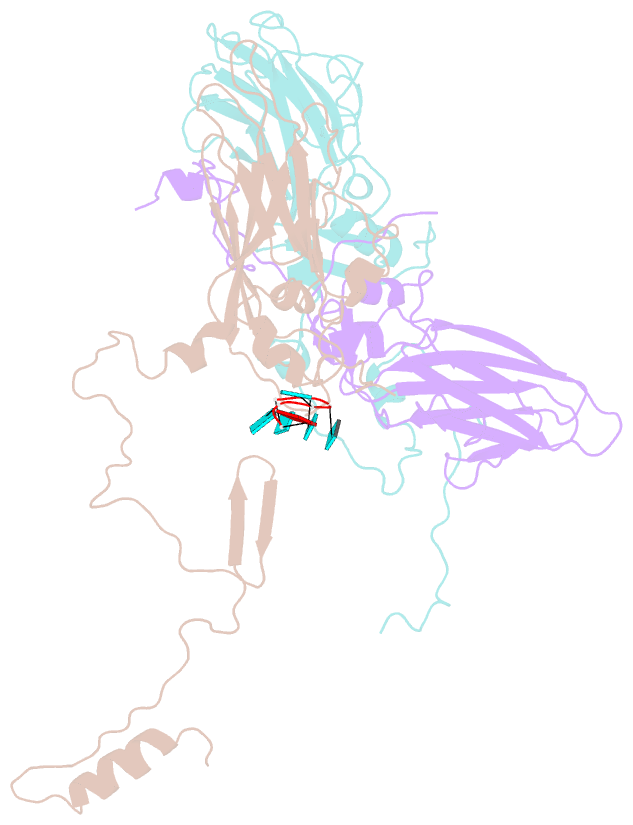
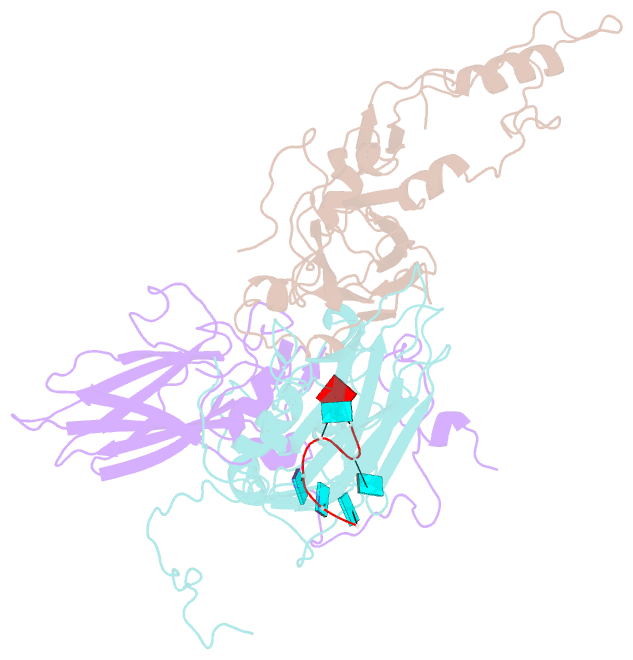
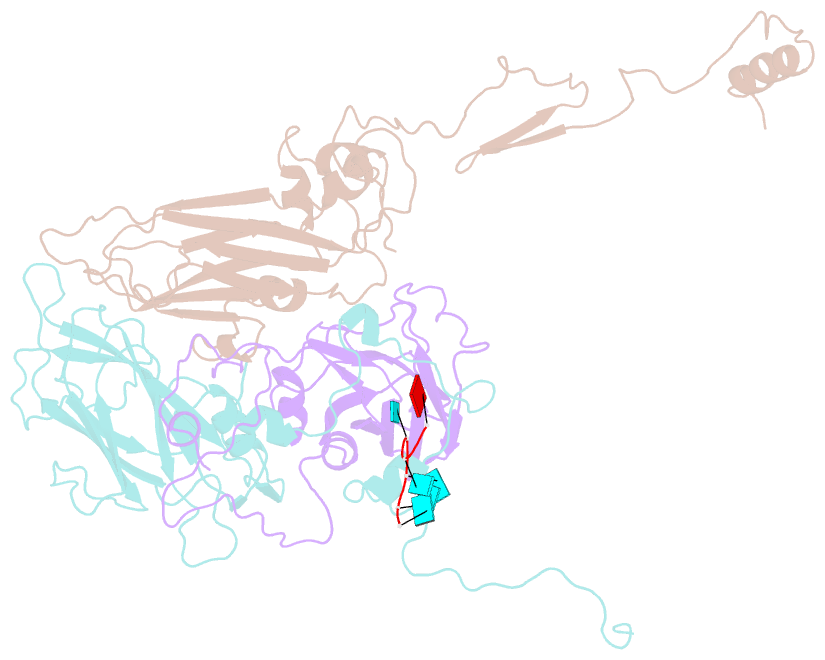
SNAP output for PDB entry 4z92 [SNAP web server]
Summary information and primary citation
- PDB-id
- 4z92; SNAP-derived features in text and JSON formats; DNAproDB
- Class
- virus
- Method
- X-ray (3.1 Å)
- Summary
- Crystal structure of parechovirus-1 virion
- Reference
- Kalynych S, Palkova L, Plevka P (2015): "The Structure of Human Parechovirus 1 Reveals an Association of the RNA Genome with the Capsid." J.Virol., 90, 1377-1386. doi: 10.1128/JVI.02346-15.
- Abstract