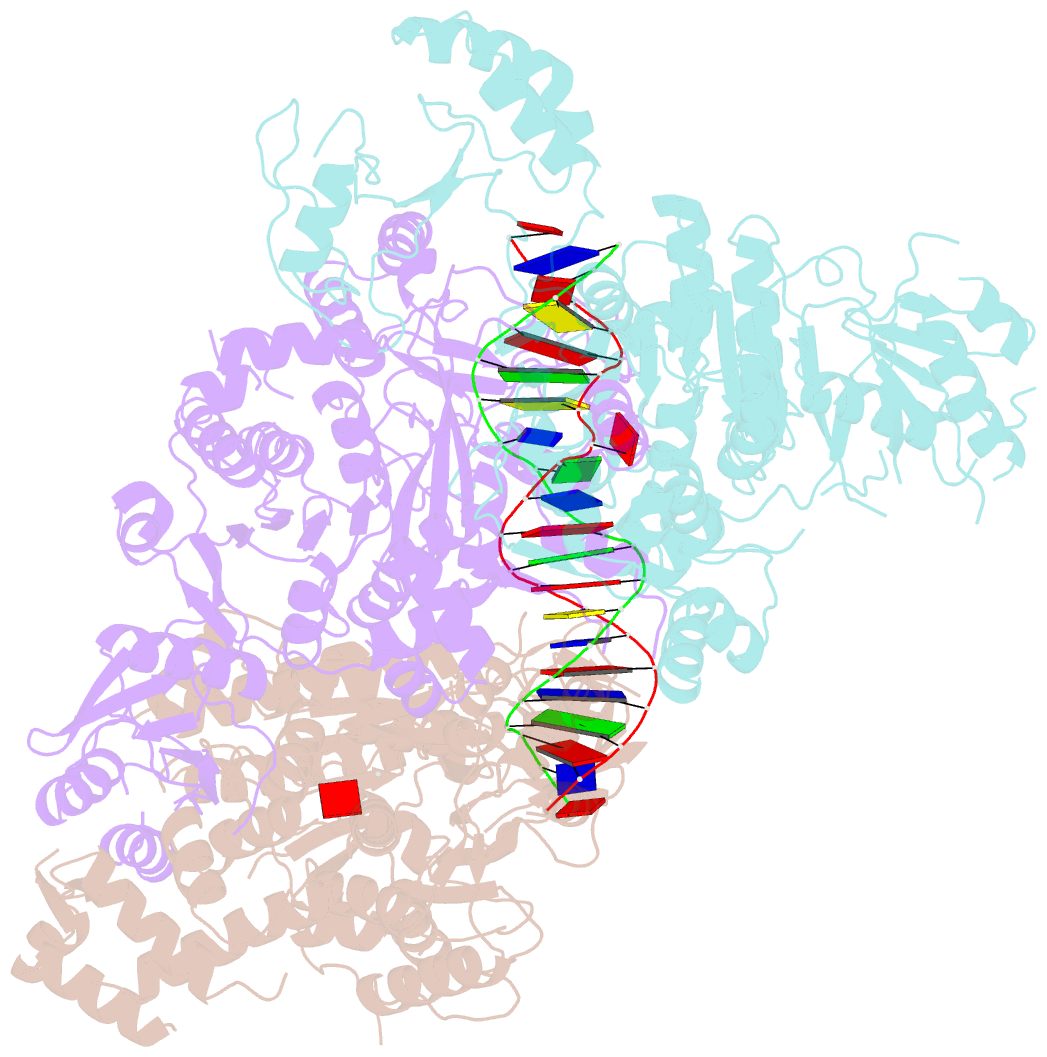
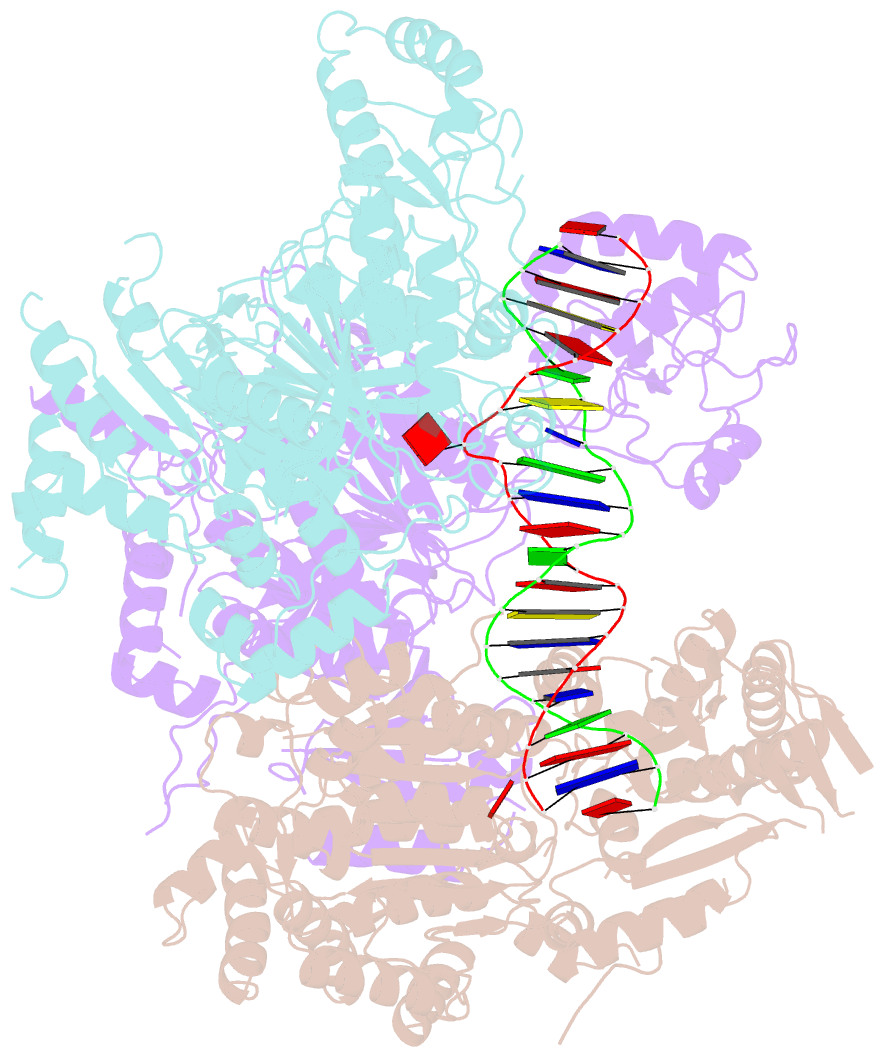
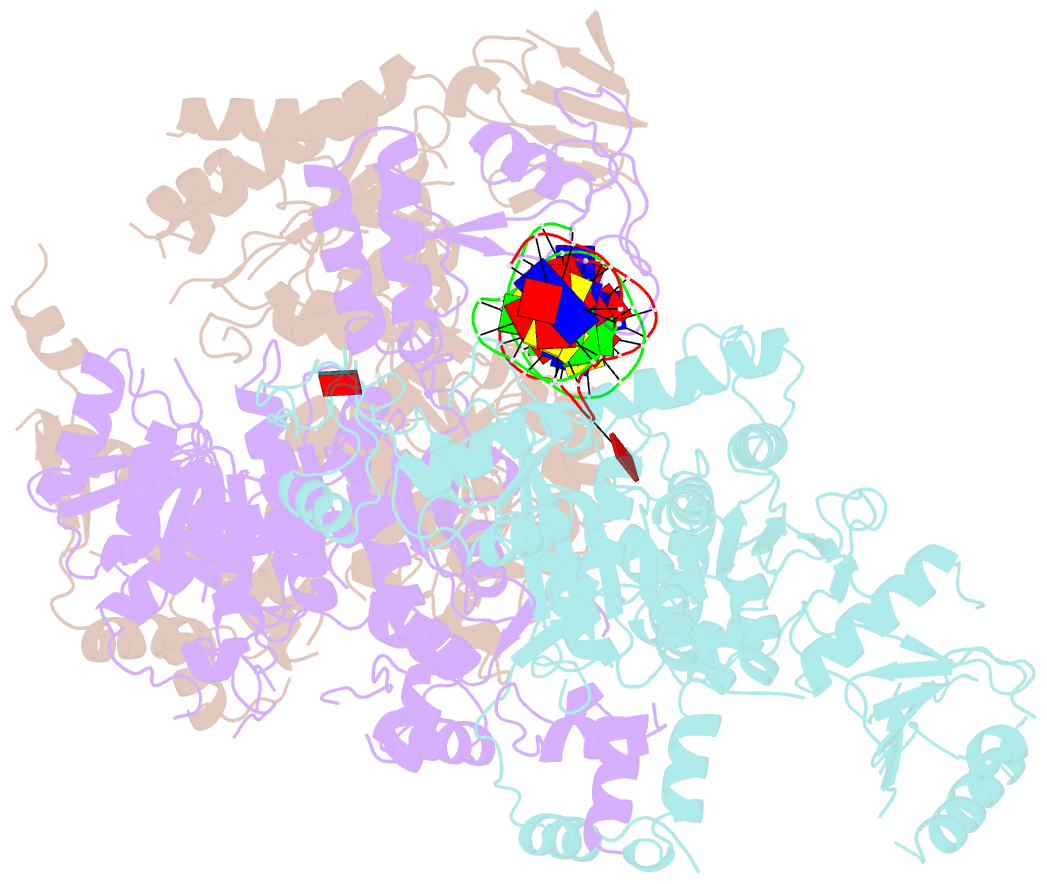
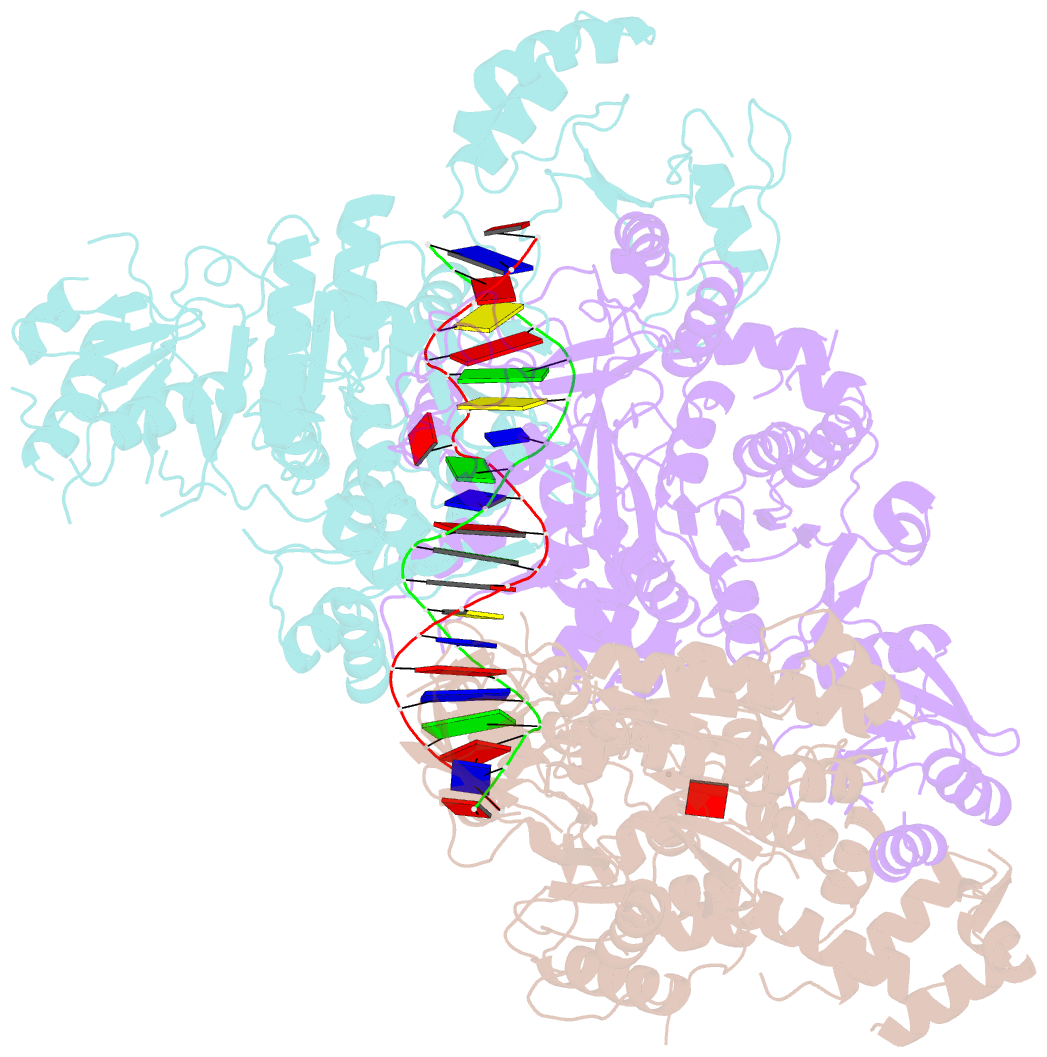
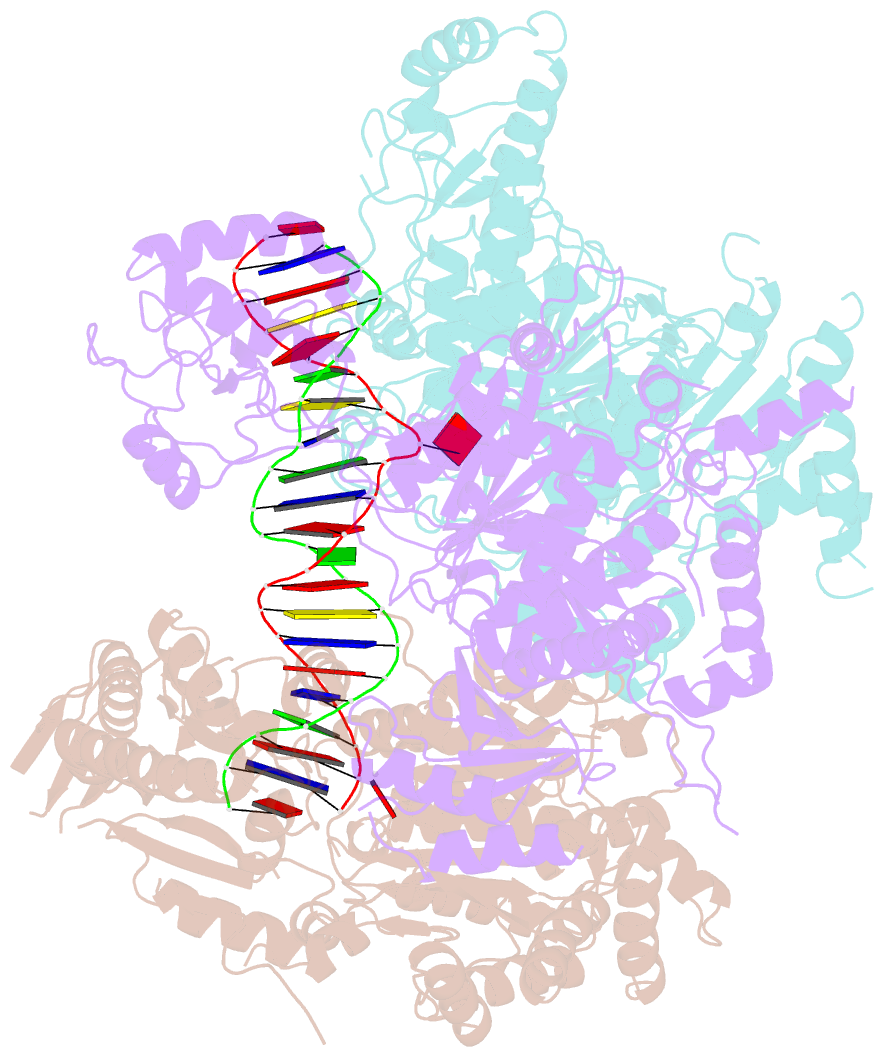
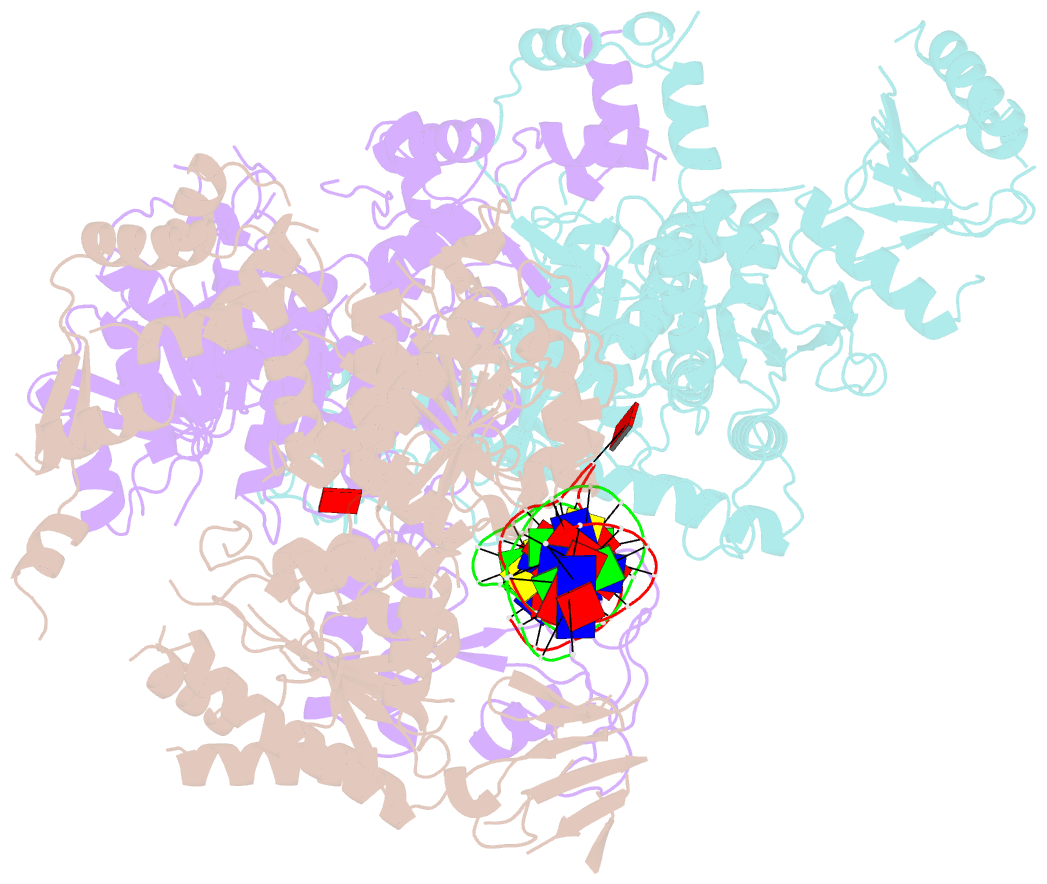
Summary information and primary citation
- PDB-id
- 4zcf; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA complex
- Method
- X-ray (2.6 Å)
- Summary
- Structural basis of asymmetric DNA methylation and atp-triggered long-range diffusion by ecop15i
- Reference
- Gupta YK, Chan SH, Xu SY, Aggarwal AK (2015): "Structural basis of asymmetric DNA methylation and ATP-triggered long-range diffusion by EcoP15I." Nat Commun, 6, 7363. doi: 10.1038/ncomms8363.
- Abstract
- Type III R-M enzymes were identified >40 years ago and yet there is no structural information on these multisubunit enzymes. Here we report the structure of a Type III R-M system, consisting of the entire EcoP15I complex (Mod2Res1) bound to DNA. The structure suggests how ATP hydrolysis is coupled to long-range diffusion of a helicase on DNA, and how a dimeric methyltransferase functions to methylate only one of the two DNA strands. We show that the EcoP15I motor domains are specifically adapted to bind double-stranded DNA and to facilitate DNA sliding via a novel 'Pin' domain. We also uncover unexpected 'division of labour', where one Mod subunit recognizes DNA, while the other Mod subunit methylates the target adenine--a mechanism that may extend to adenine N6 RNA methylation in mammalian cells. Together the structure sheds new light on the mechanisms of both helicases and methyltransferases in DNA and RNA metabolism.