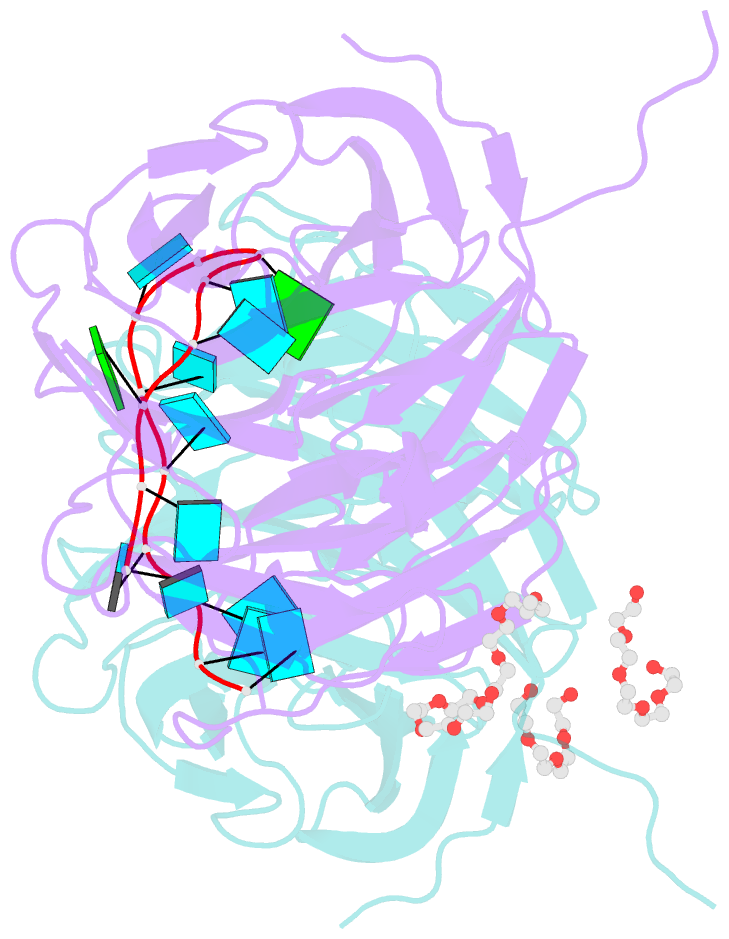
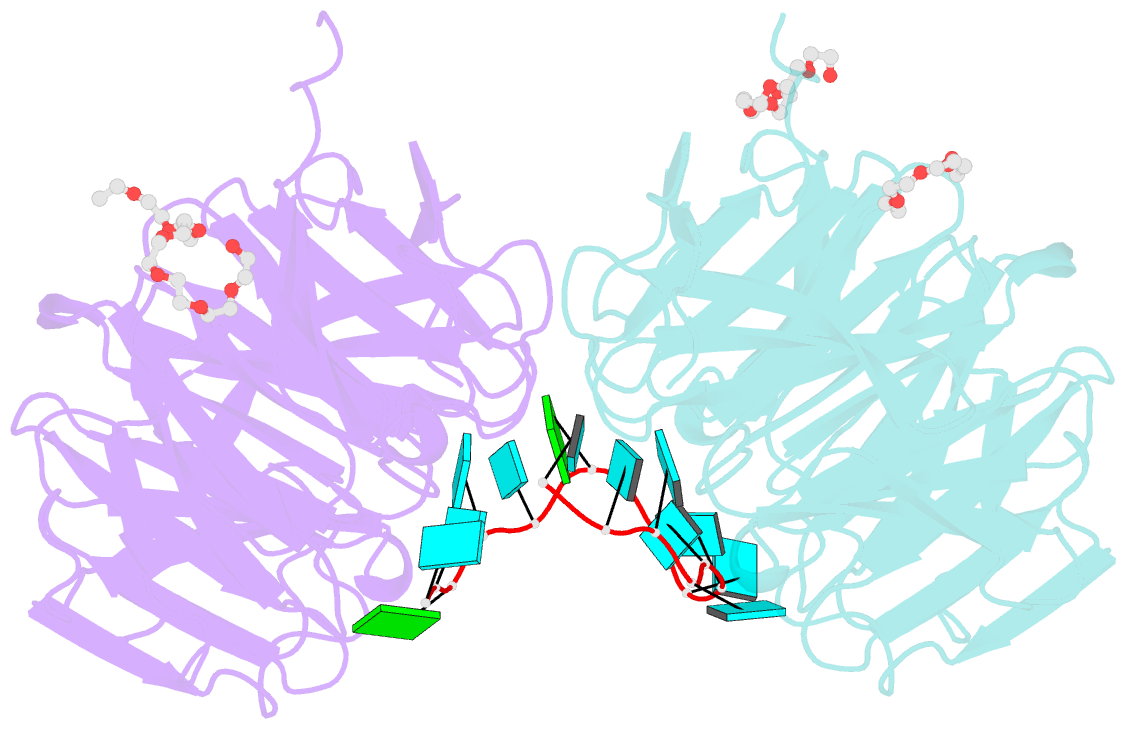
Summary information and primary citation
- PDB-id
- 4zlr; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- translation
- Method
- X-ray (2.3 Å)
- Summary
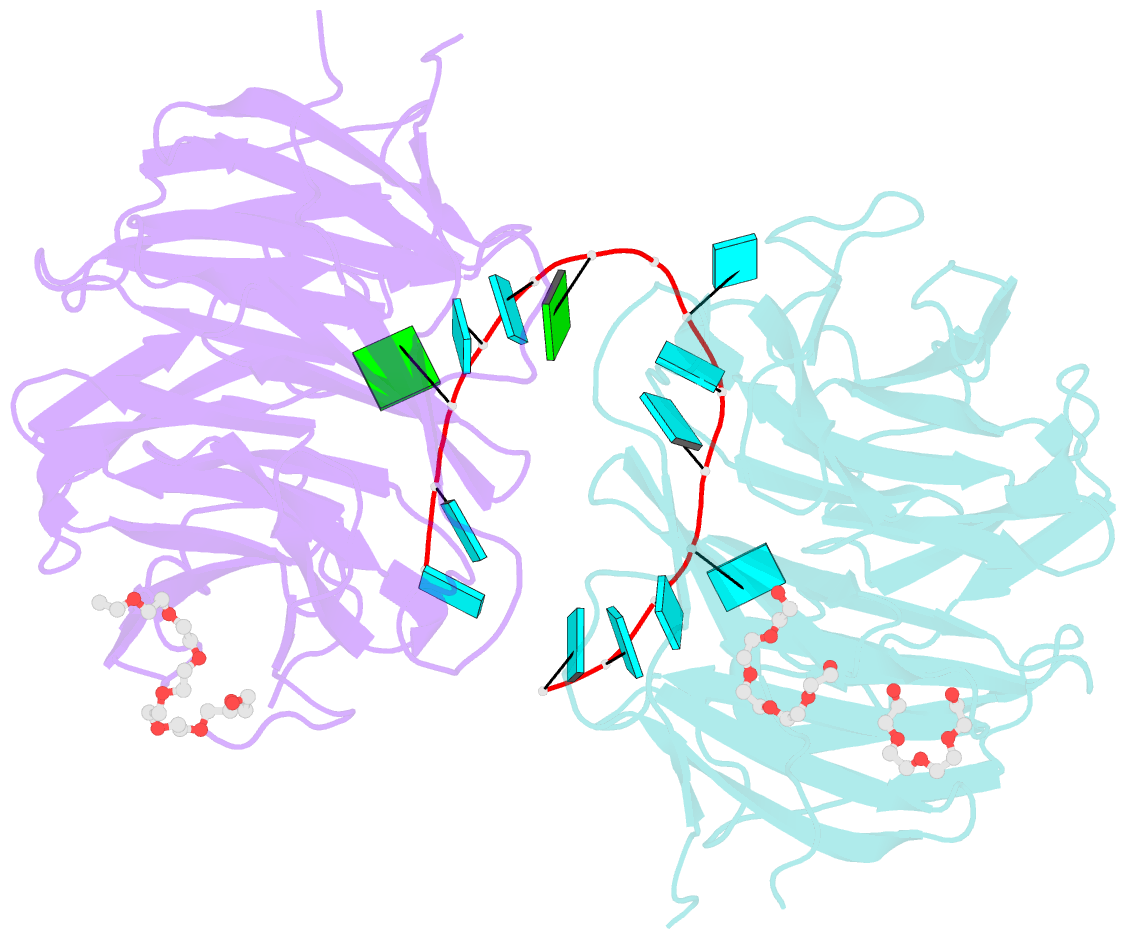
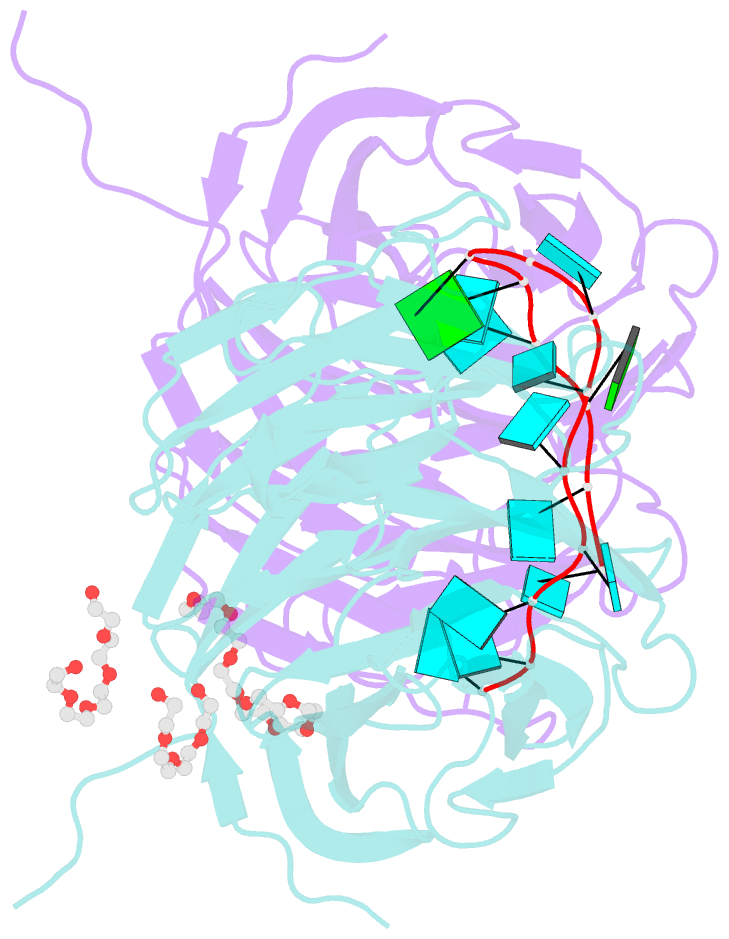
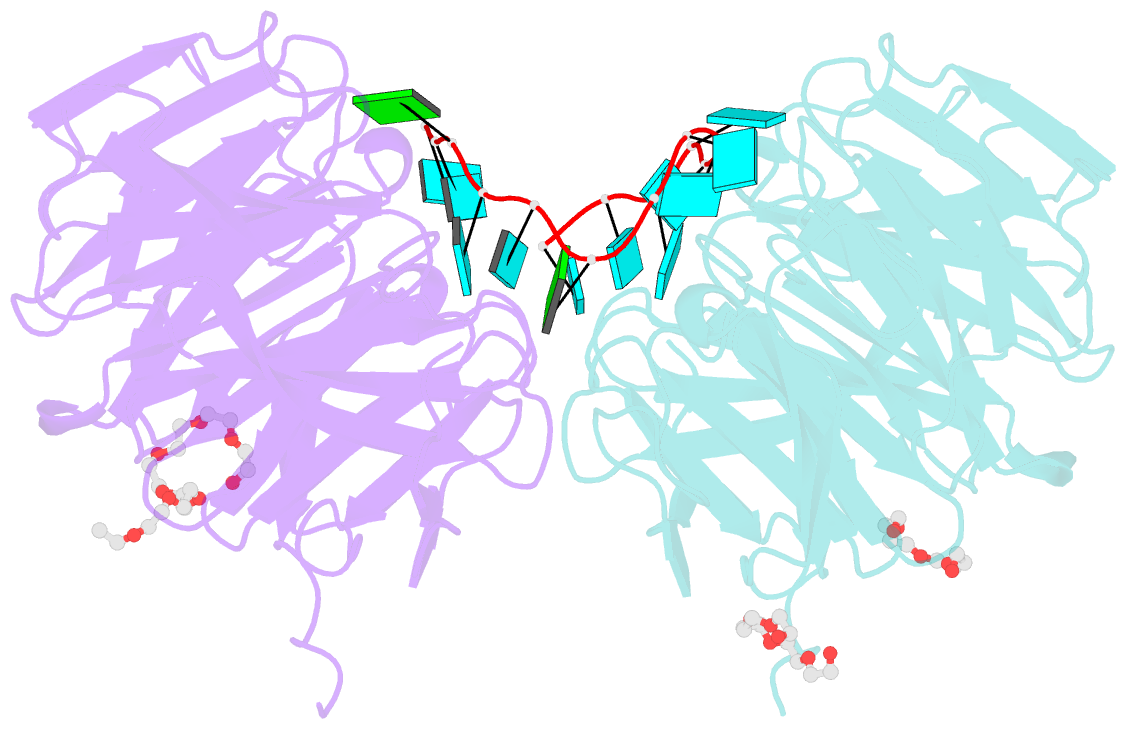
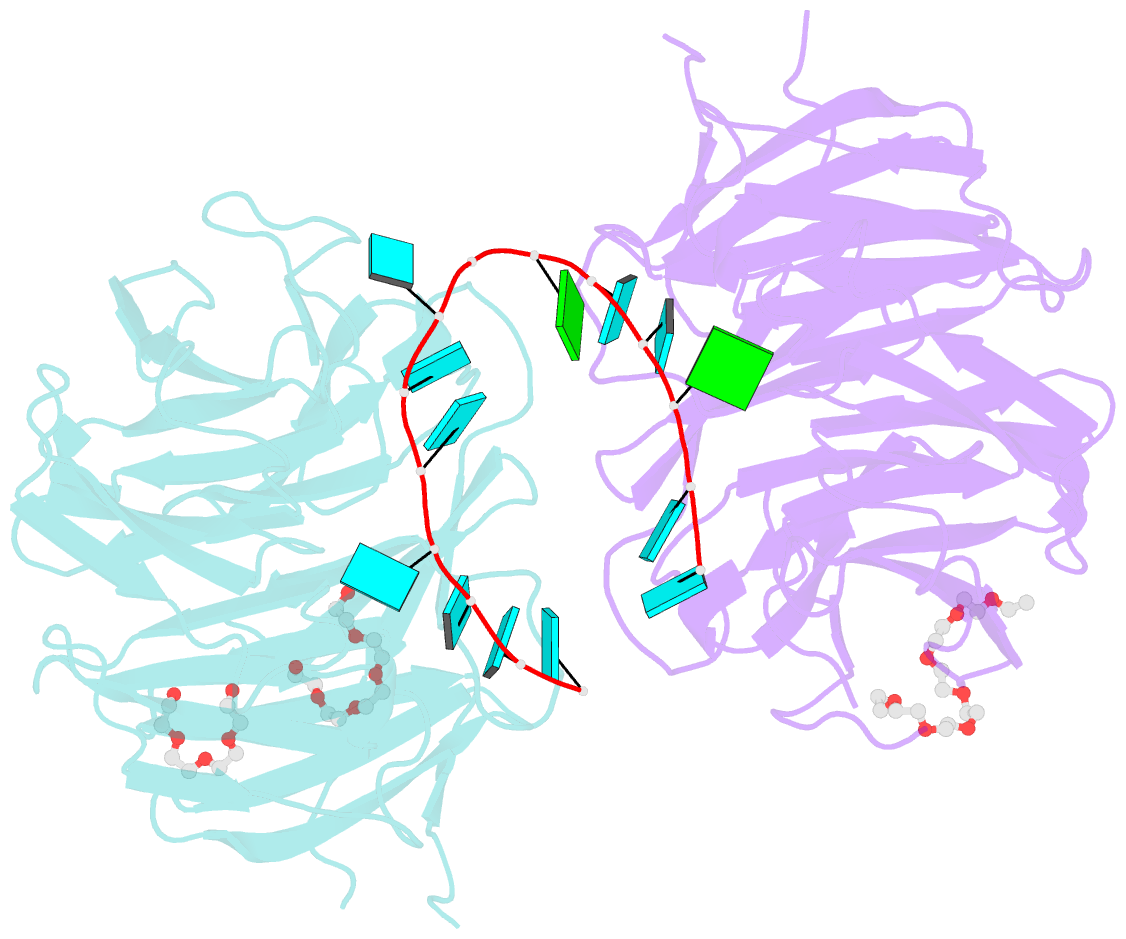
- Structure of the brat-nhl domain bound to consensus RNA motif
- Reference
- Loedige I, Jakob L, Treiber T, Ray D, Stotz M, Treiber N, Hennig J, Cook KB, Morris Q, Hughes TR, Engelmann JC, Krahn MP, Meister G (2015): "The Crystal Structure of the NHL Domain in Complex with RNA Reveals the Molecular Basis of Drosophila Brain-Tumor-Mediated Gene Regulation." Cell Rep, 13, 1206-1220. doi: 10.1016/j.celrep.2015.09.068.
- Abstract
- TRIM-NHL proteins are conserved among metazoans and control cell fate decisions in various stem cell linages. The Drosophila TRIM-NHL protein Brain tumor (Brat) directs differentiation of neuronal stem cells by suppressing self-renewal factors. Brat is an RNA-binding protein and functions as a translational repressor. However, it is unknown which RNAs Brat regulates and how RNA-binding specificity is achieved. Using RNA immunoprecipitation and RNAcompete, we identify Brat-bound mRNAs in Drosophila embryos and define consensus binding motifs for Brat as well as a number of additional TRIM-NHL proteins, indicating that TRIM-NHL proteins are conserved, sequence-specific RNA-binding proteins. We demonstrate that Brat-mediated repression and direct RNA-binding depend on the identified motif and show that binding of the localization factor Miranda to the Brat-NHL domain inhibits Brat activity. Finally, to unravel the sequence specificity of the NHL domain, we crystallize the Brat-NHL domain in complex with RNA and present a high-resolution protein-RNA structure of this fold.