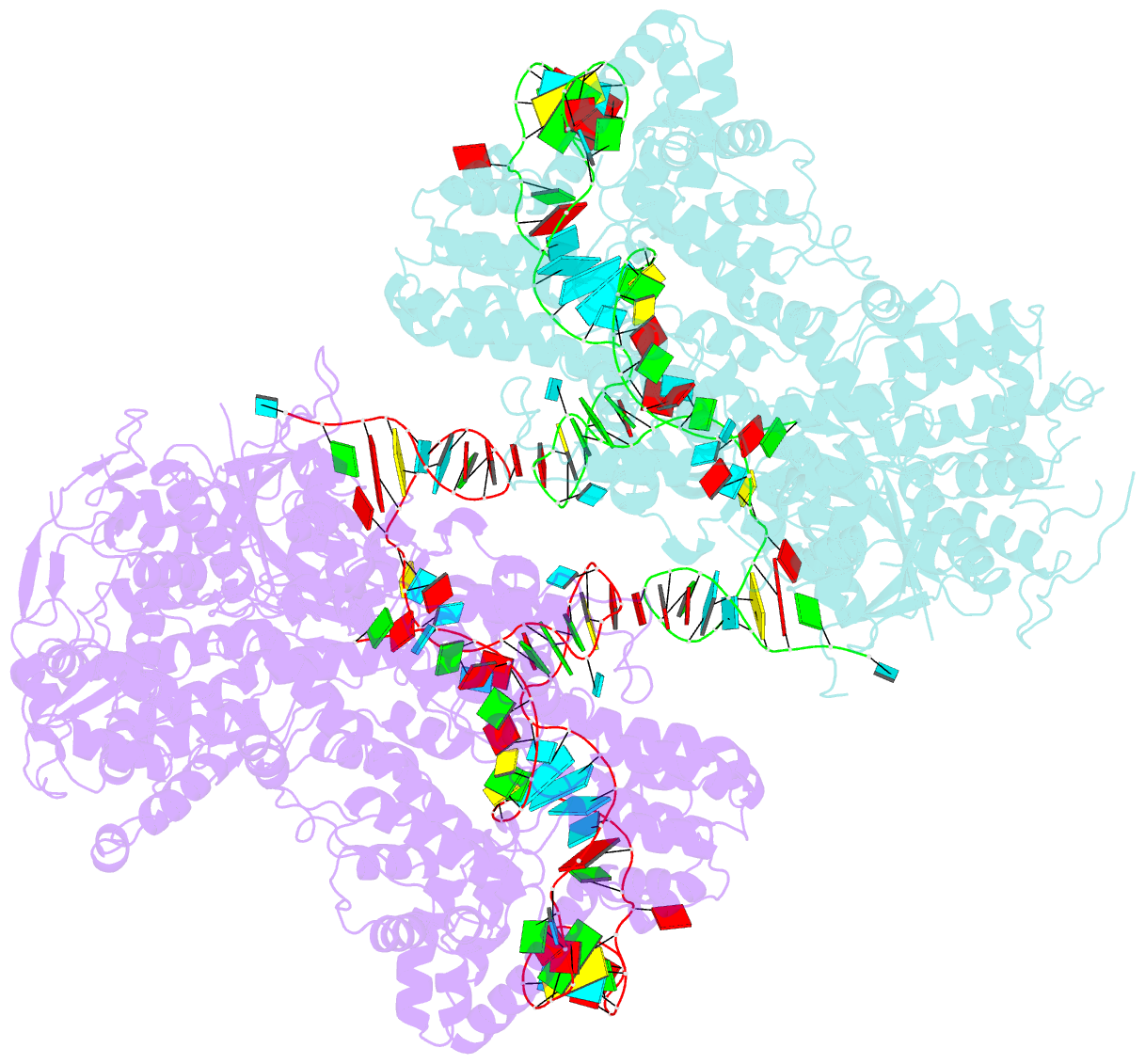
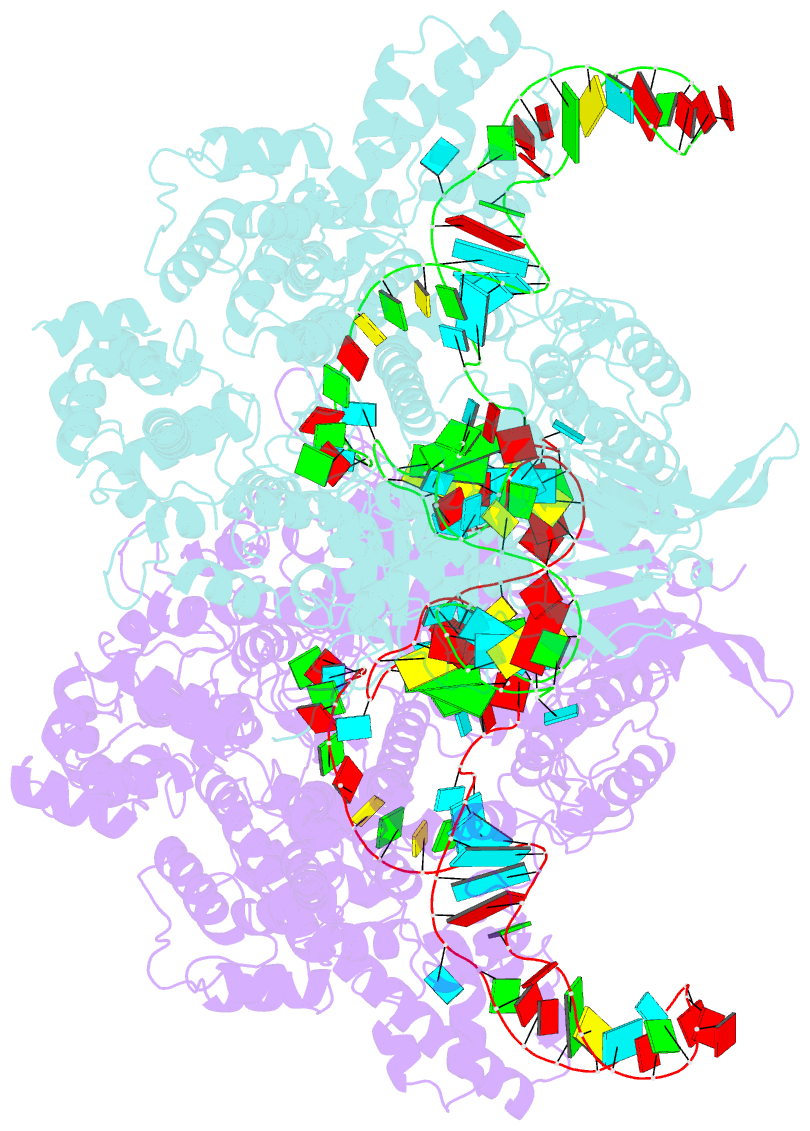
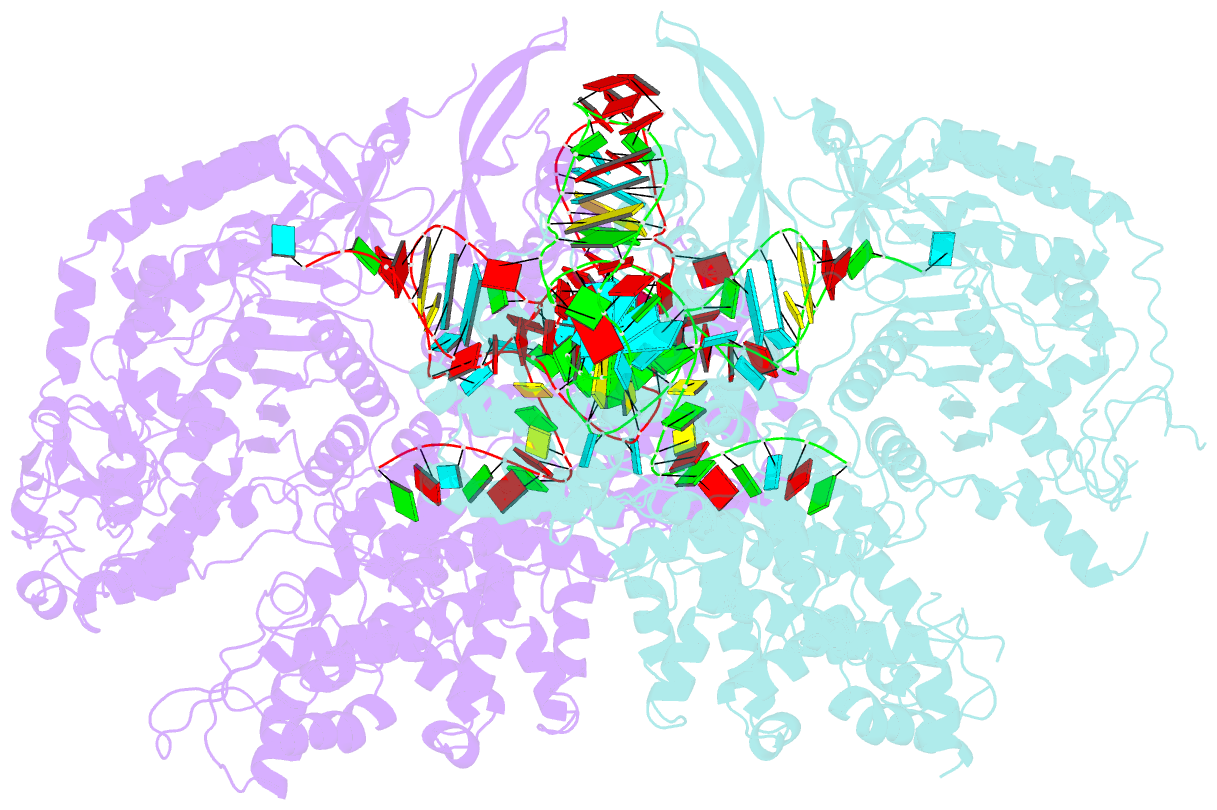
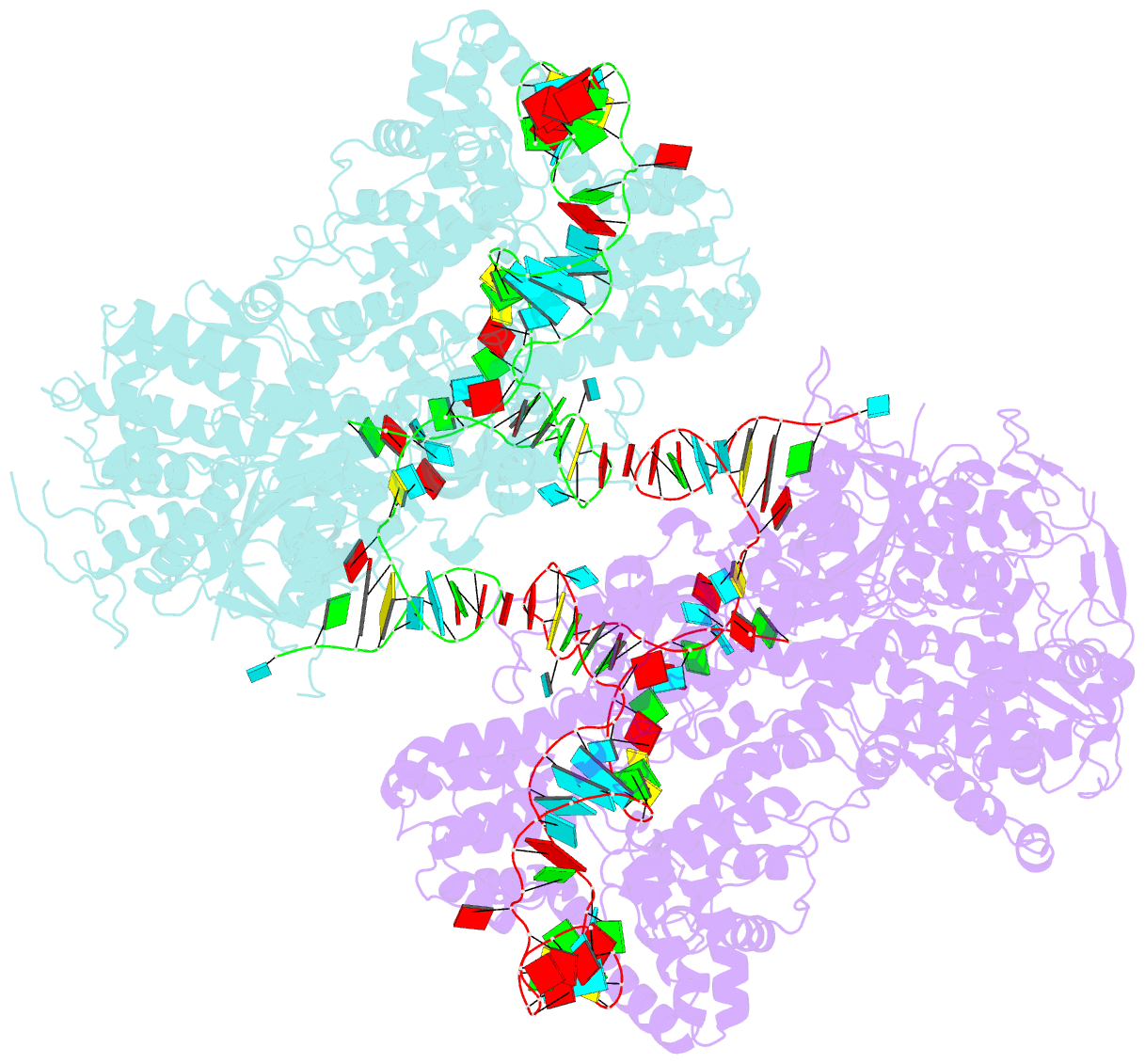
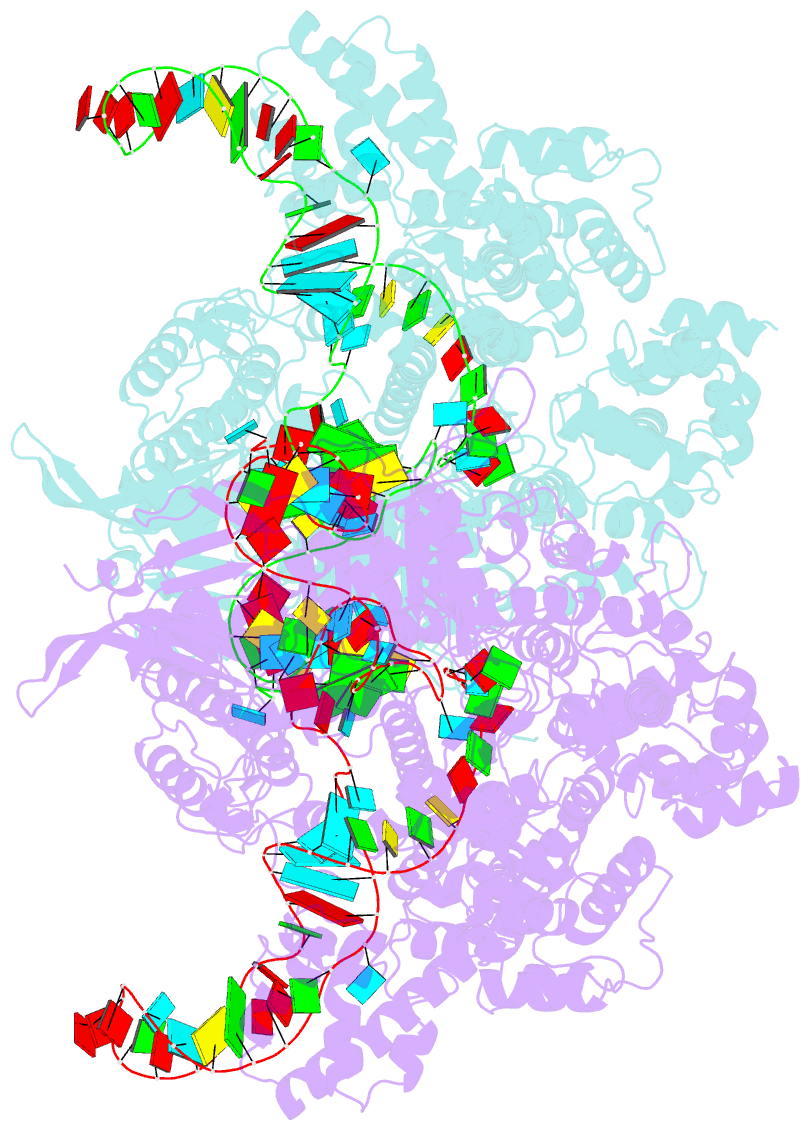
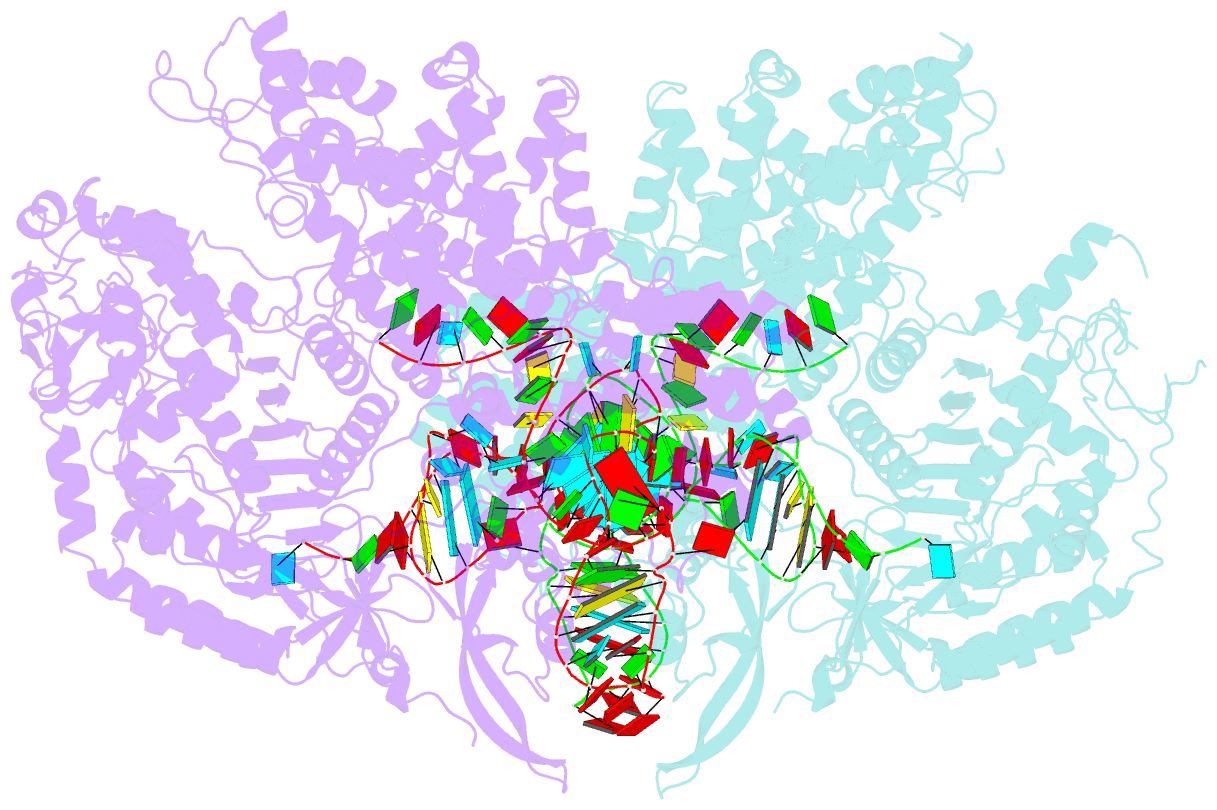
Summary information and primary citation
- PDB-id
- 4zt9; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (3.1 Å)
- Summary
- Nuclease-inactive streptococcus pyogenes cas9 (d10a-h840a, dcas9) in complex with single-guide RNA at 3.1 angstrom resolution
- Reference
- Jiang F, Zhou K, Ma L, Gressel S, Doudna JA (2015): "A Cas9-guide RNA complex preorganized for target DNA recognition." Science, 348, 1477-1481. doi: 10.1126/science.aab1452.
- Abstract
- Bacterial adaptive immunity uses CRISPR (clustered regularly interspaced short palindromic repeats)-associated (Cas) proteins together with CRISPR transcripts for foreign DNA degradation. In type II CRISPR-Cas systems, activation of Cas9 endonuclease for DNA recognition upon guide RNA binding occurs by an unknown mechanism. Crystal structures of Cas9 bound to single-guide RNA reveal a conformation distinct from both the apo and DNA-bound states, in which the 10-nucleotide RNA "seed" sequence required for initial DNA interrogation is preordered in an A-form conformation. This segment of the guide RNA is essential for Cas9 to form a DNA recognition-competent structure that is poised to engage double-stranded DNA target sequences. We construe this as convergent evolution of a "seed" mechanism reminiscent of that used by Argonaute proteins during RNA interference in eukaryotes.