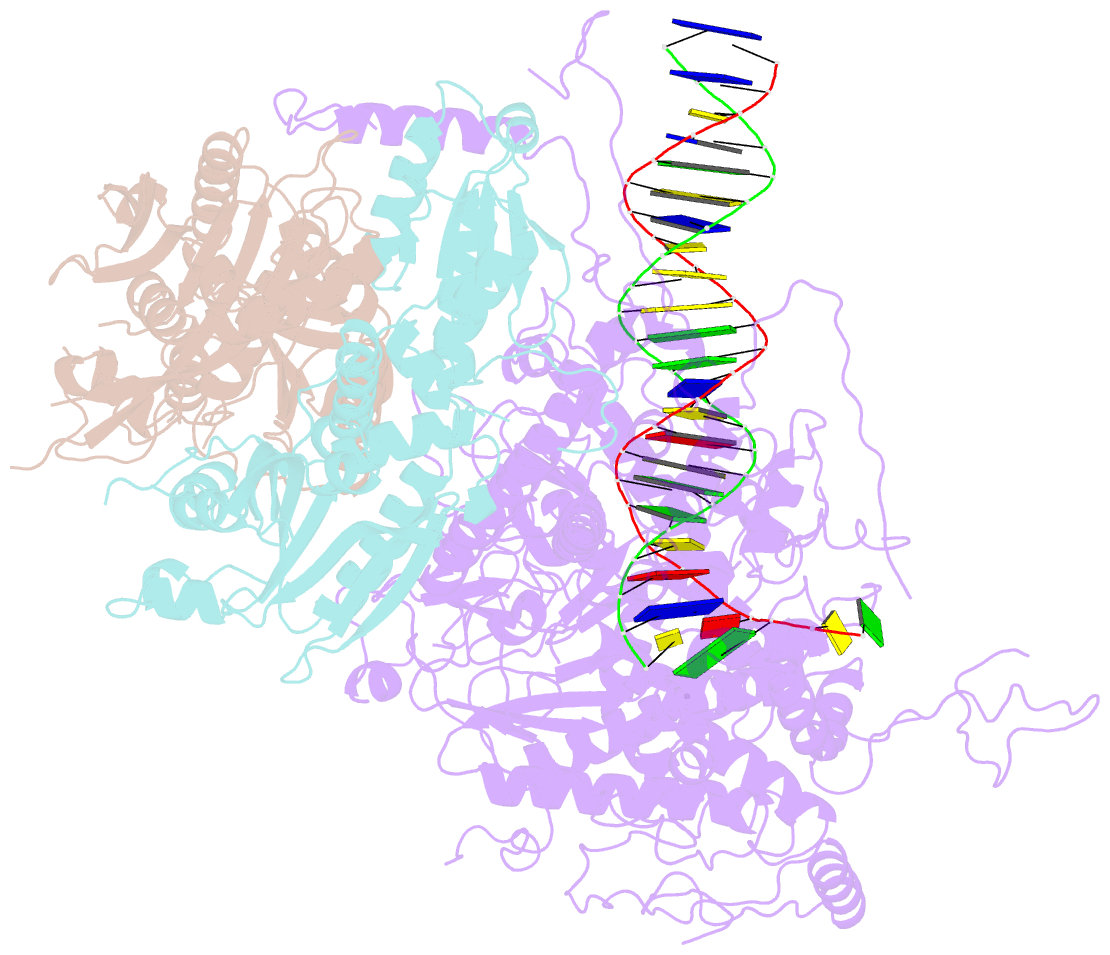
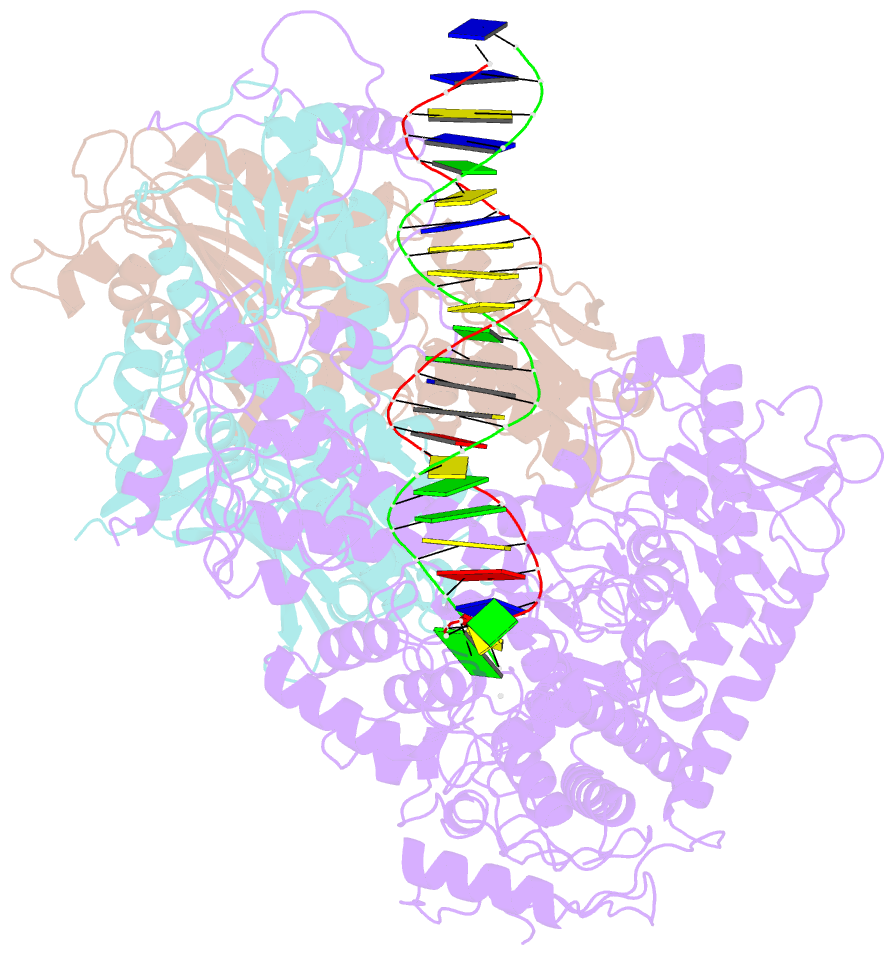
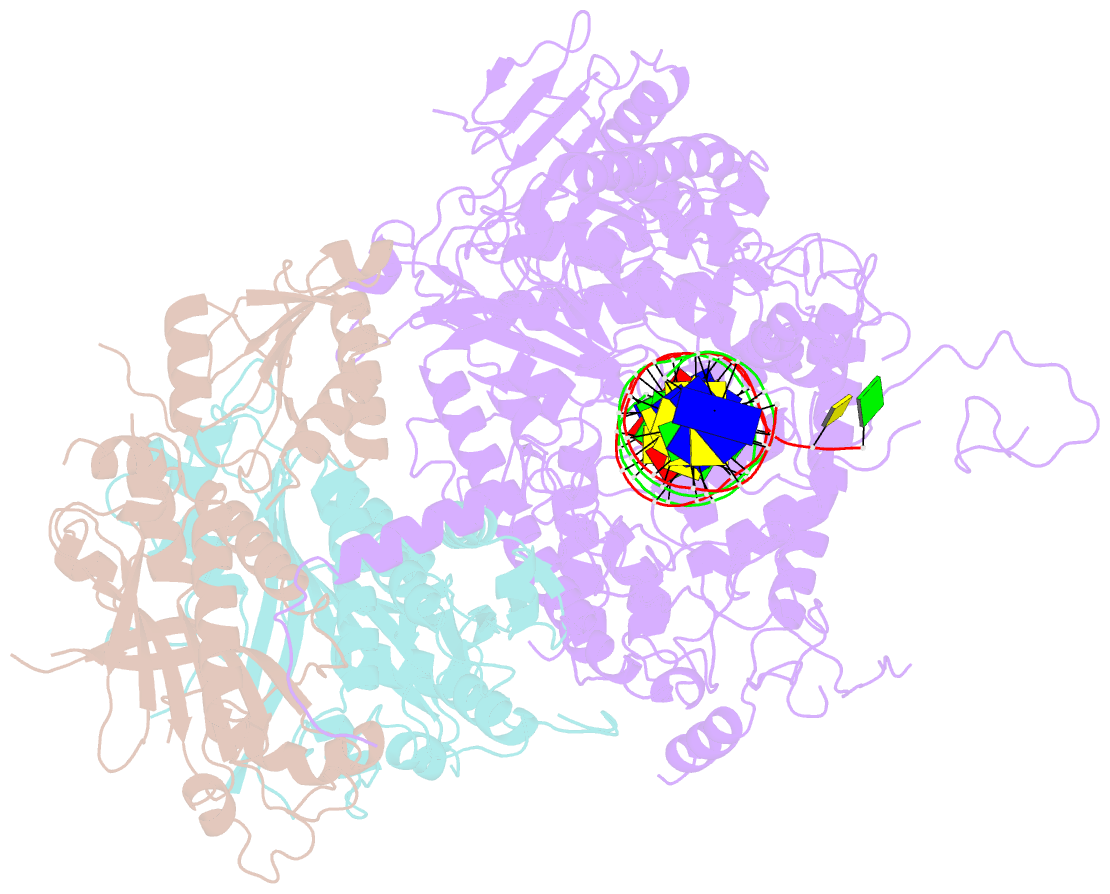
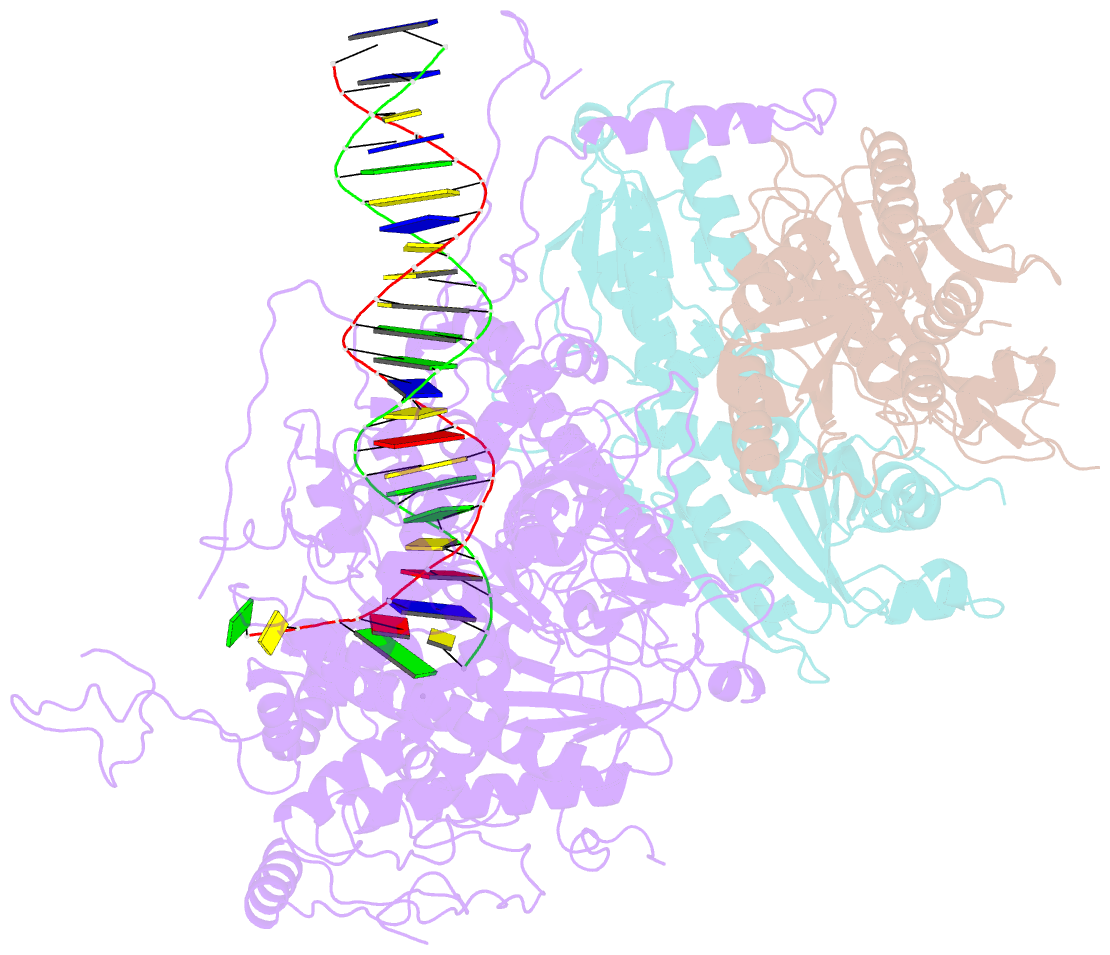
Summary information and primary citation
- PDB-id
- 4ztz; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (3.442 Å)
- Summary
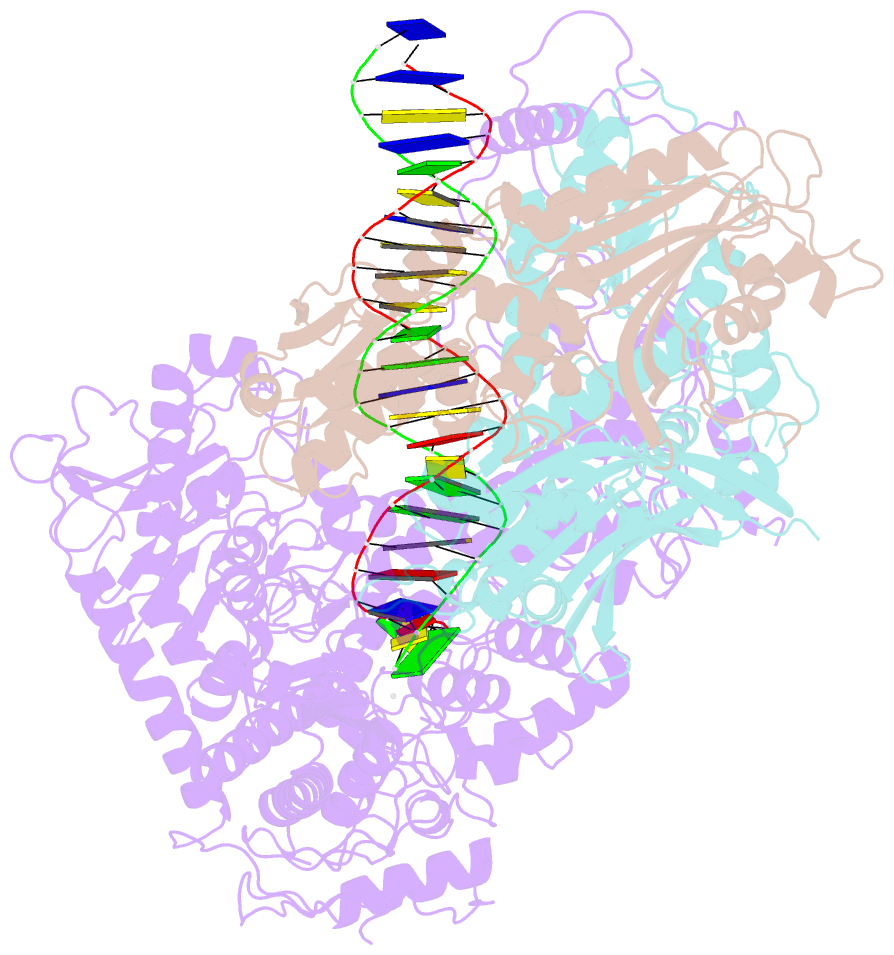
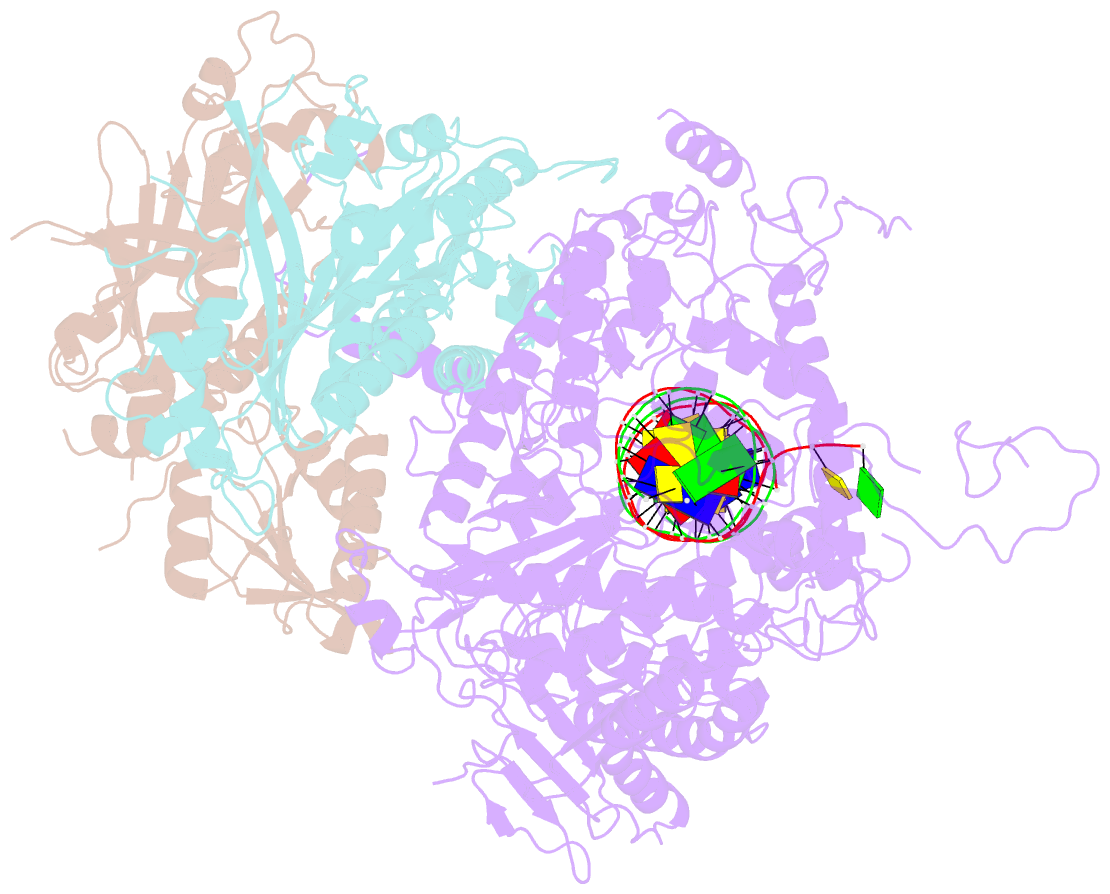
- Structural basis for processivity and antiviral drug toxicity in human mitochondrial DNA replicase
- Reference
- Szymanski MR, Kuznestov VB, Shumate CK, Meng Q, Lee Y-S, Patel G, Patel SS, Yin YW (2015): "Structural basis for processivity and antiviral drug toxicity
in human mitochondrial DNA replicase." EMBO J., 34, 1959. doi: 10.15252/embj.201591520.
- Abstract
- The human DNA polymerase gamma (Pol γ) is responsible for DNA replication in mitochondria. Pol γ is particularly susceptible to inhibition by dideoxynucleoside-based inhibitors designed to fight viral infection. Here, we report crystal structures of the replicating Pol γ-DNA complex bound to either substrate or zalcitabine, an inhibitor used for HIV reverse transcriptase. The structures reveal that zalcitabine binds to the Pol γ active site almost identically to the substrate dCTP, providing a structural basis for Pol γ-mediated drug toxicity. When compared to the apo form, Pol γ undergoes intra- and inter-subunit conformational changes upon formation of the ternary complex with primer/template DNA and substrate. We also find that the accessory subunit Pol γB, which lacks intrinsic enzymatic activity and does not contact the primer/template DNA directly, serves as an allosteric regulator of holoenzyme activities. The structures presented here suggest a mechanism for processivity of the holoenzyme and provide a model for understanding the deleterious effects of Pol γ mutations in human disease. Crystal structures of the mitochondrial DNA polymerase, Pol γ, in complex with substrate or antiviral inhibitor zalcitabine provide a basis for understanding Pol γ-mediated drug toxicity.