Summary information and primary citation
- PDB-id
- 5a0m; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-DNA
- Method
- X-ray (2.9 Å)
- Summary
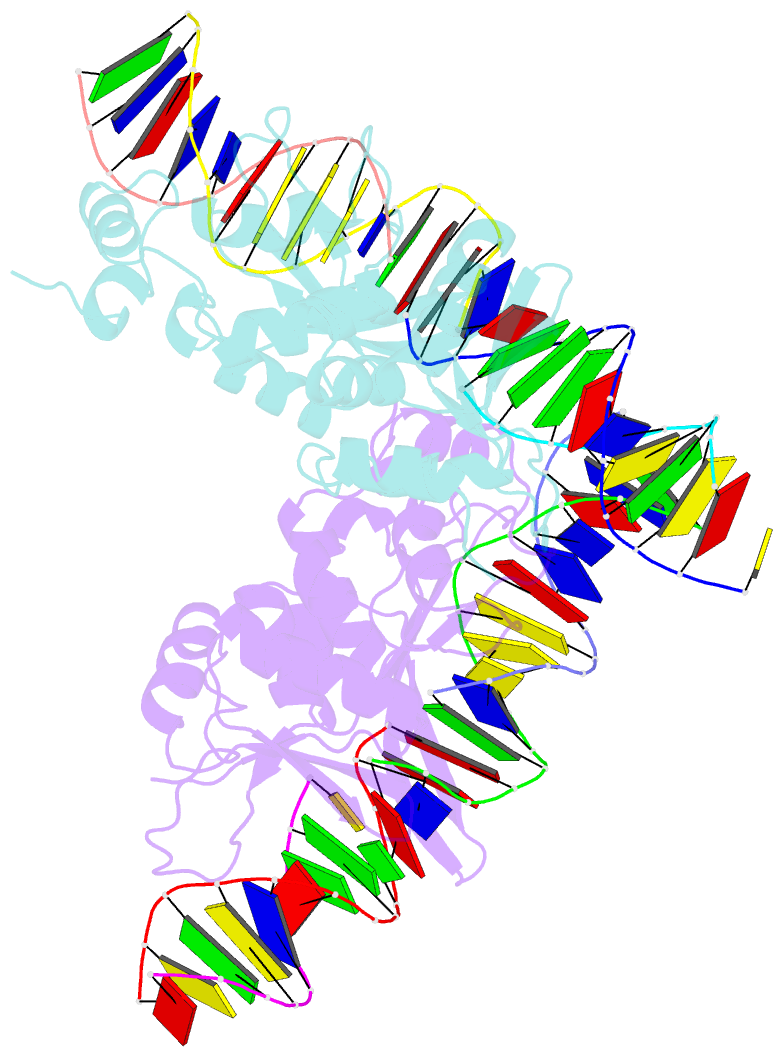
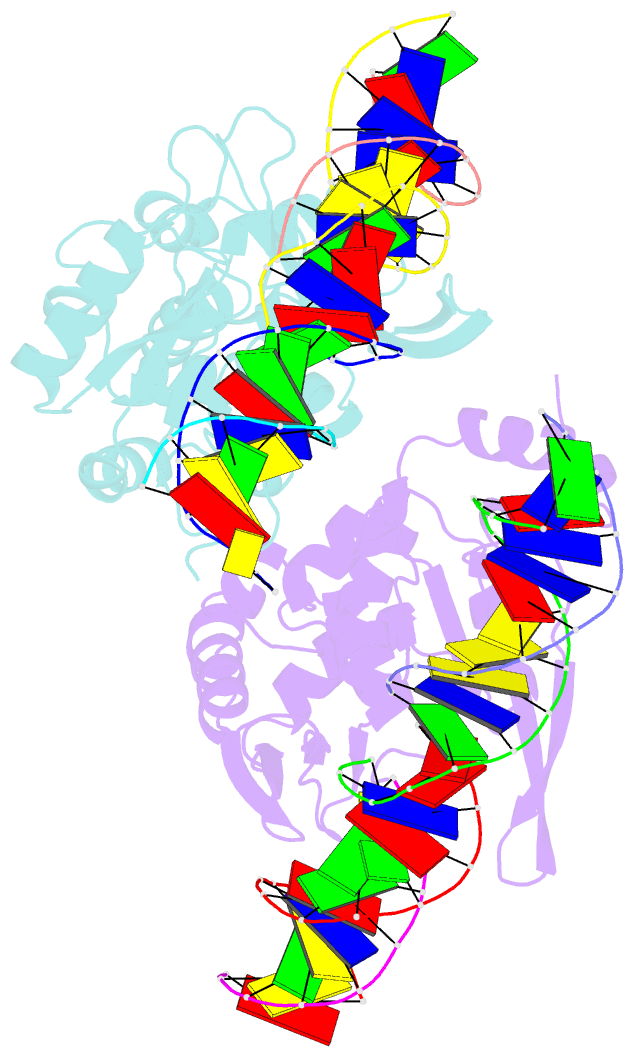
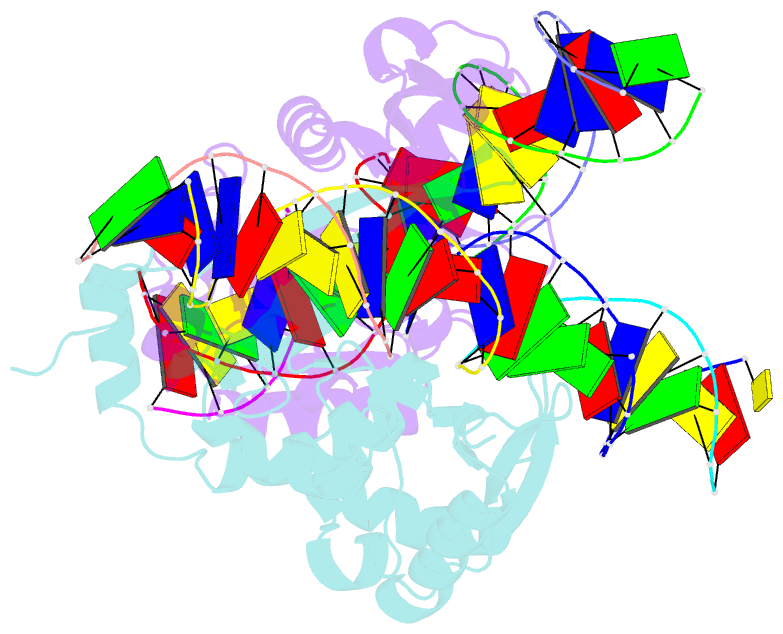
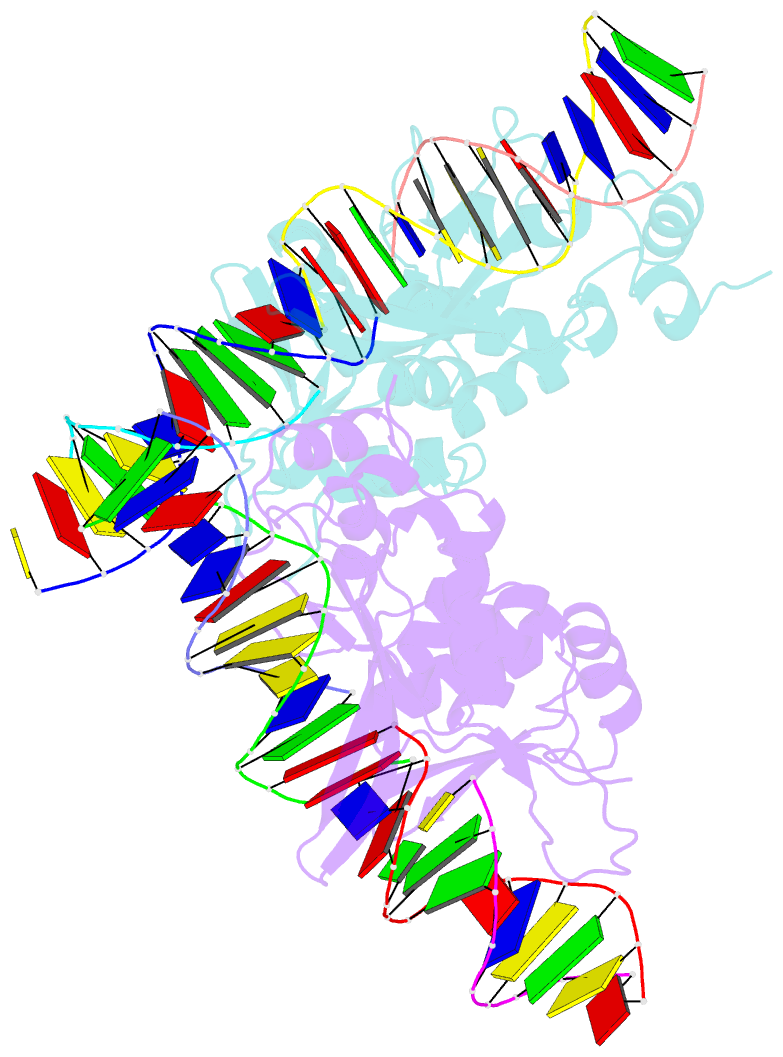
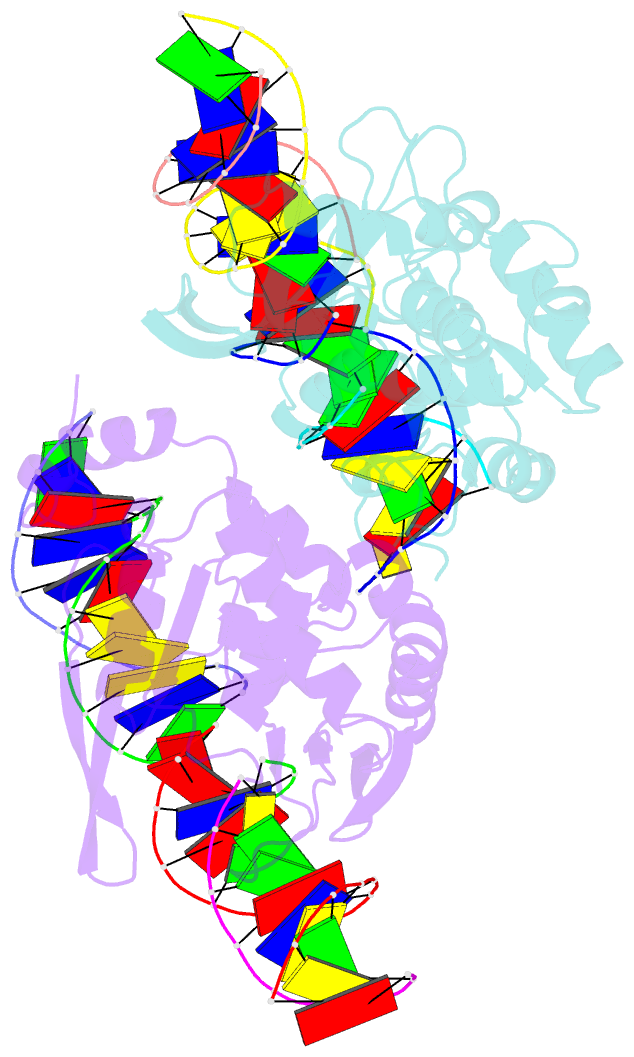
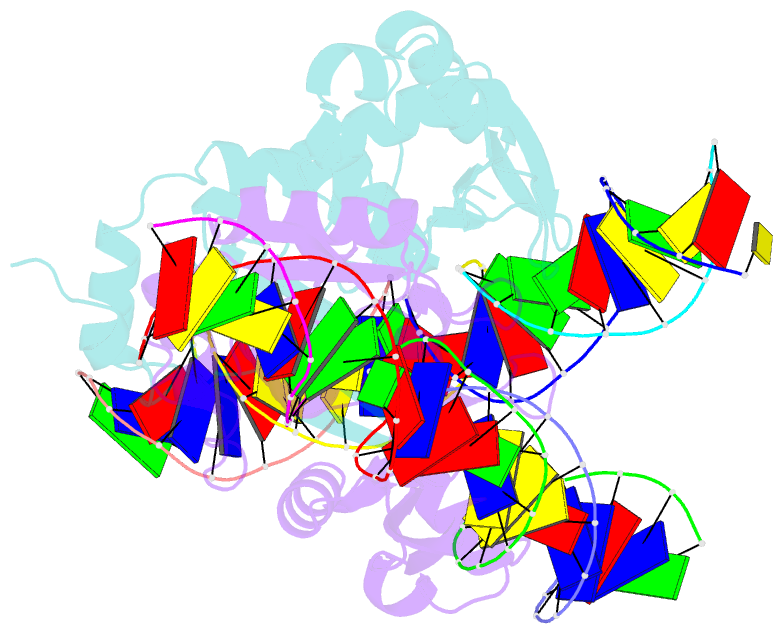
- The crystal structure of i-scei in complex with its target DNA in the presence of mn
- Reference
- Prieto J, Redondo P, Merino N, Villate M, Montoya G, Blanco FJ, Molina R (2016): "Structure of the I-Scei Nuclease Complexed with its DsDNA Target and Three Catalytic Metal Ions." Acta Crystallogr.,Sect.F, 72, 473. doi: 10.1107/S2053230X16007512.
- Abstract
- Homing endonucleases are highly specific DNA-cleaving enzymes that recognize and cleave long stretches of DNA. The engineering of these enzymes provides instruments for genome modification in a wide range of fields, including gene targeting. The homing endonuclease I-SceI from the yeast Saccharomyces cerevisiae has been purified after overexpression in Escherichia coli and its crystal structure has been determined in complex with its target DNA. In order to evaluate the number of ions that are involved in the cleavage process, thus determining the catalytic mechanism, crystallization experiments were performed in the presence of Mn(2+), yielding crystals that were suitable for X-ray diffraction analysis. The crystals belonged to the orthorhombic space group P212121, with unit-cell parameters a = 80.11, b = 80.57, c = 130.87 Å, α = β = γ = 90°. The self-rotation function and the Matthews coefficient suggested the presence of two protein-DNA complexes in the asymmetric unit. The crystals diffracted to a resolution limit of 2.9 Å using synchrotron radiation. From the anomalous data, it was determined that three cations are involved in catalysis and it was confirmed that I-SceI follows a two-metal-ion DNA-strand cleavage mechanism.