Summary information and primary citation
- PDB-id
- 5amq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase
- Method
- X-ray (3.0 Å)
- Summary
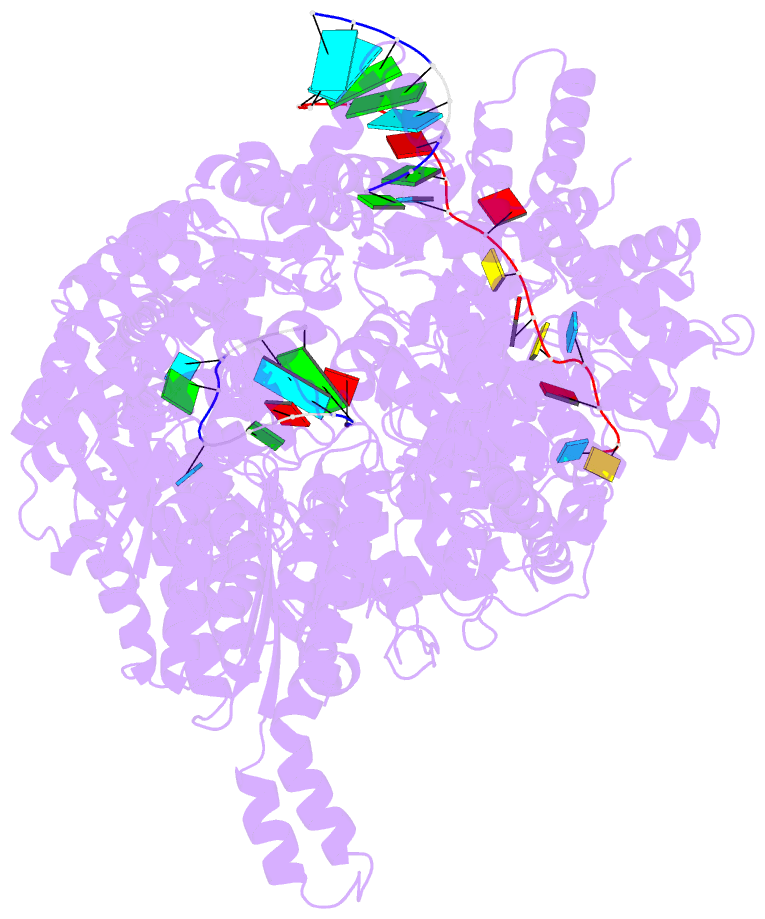
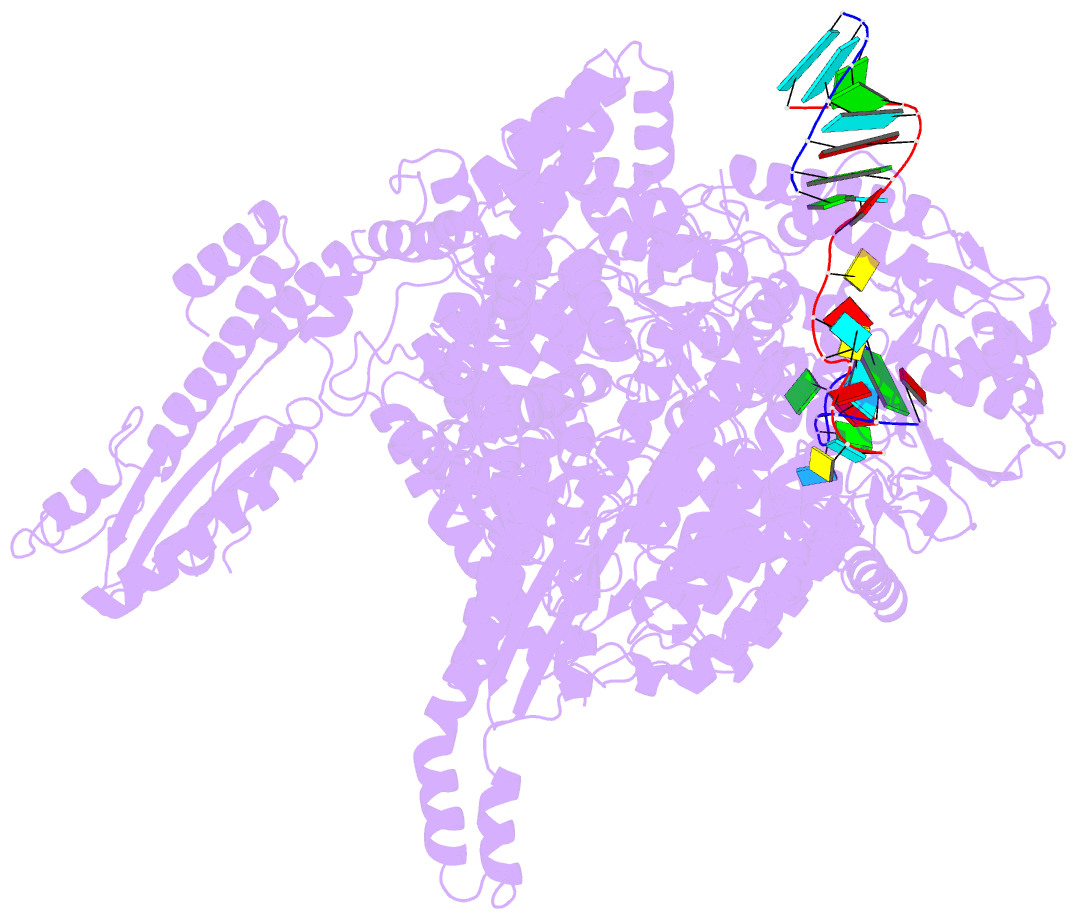
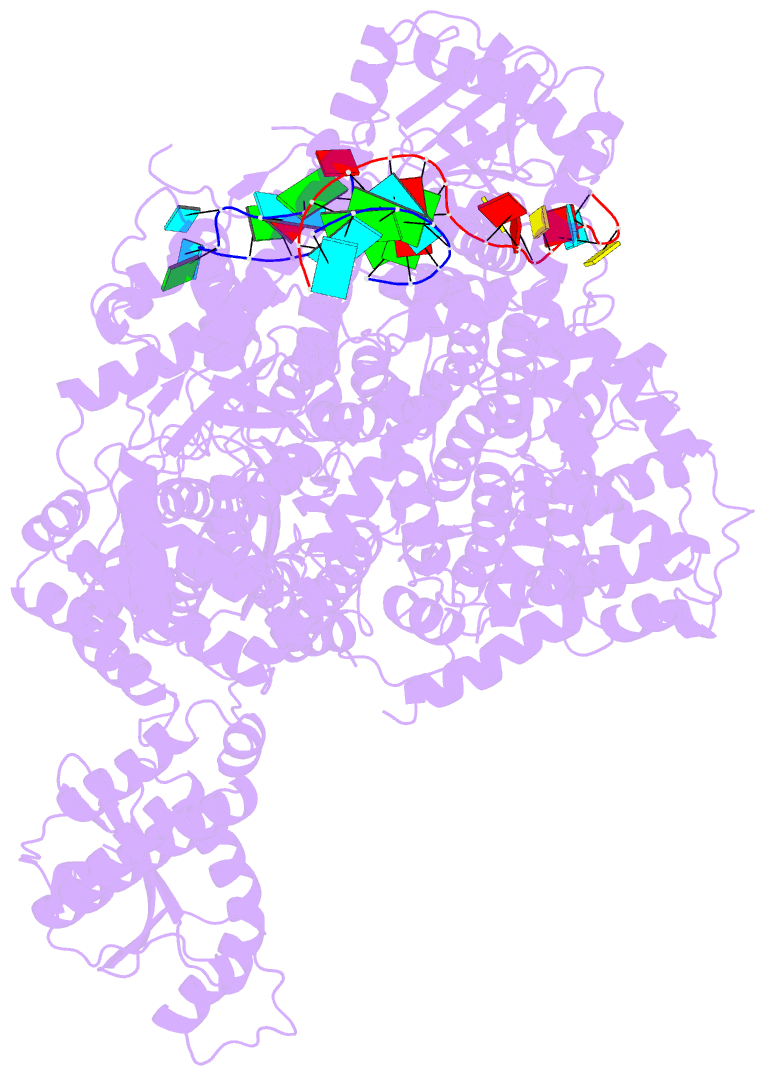
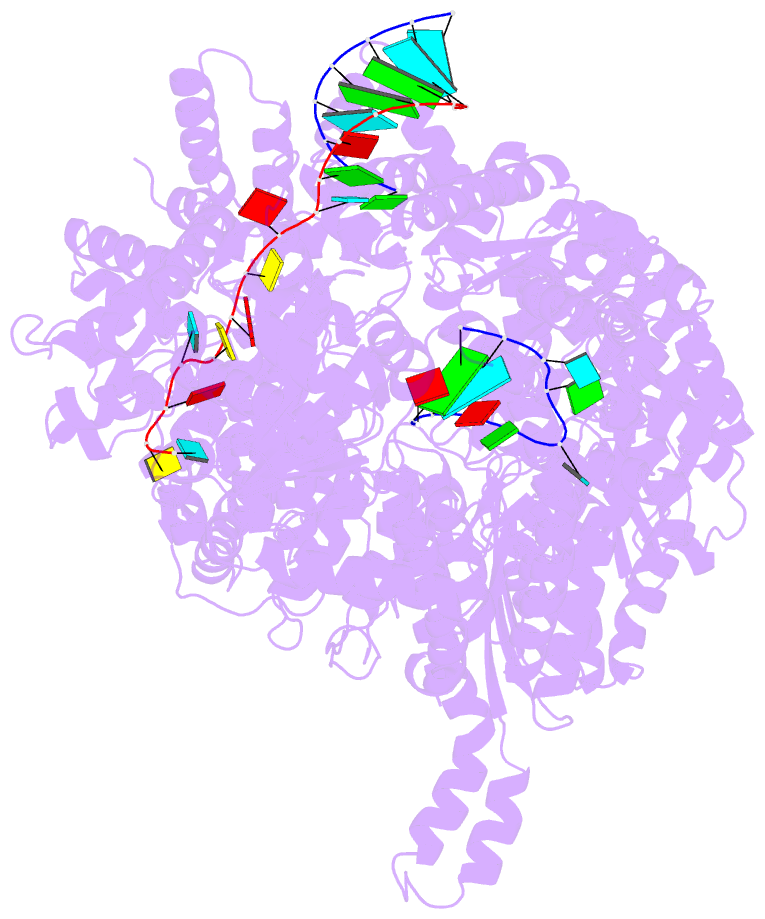
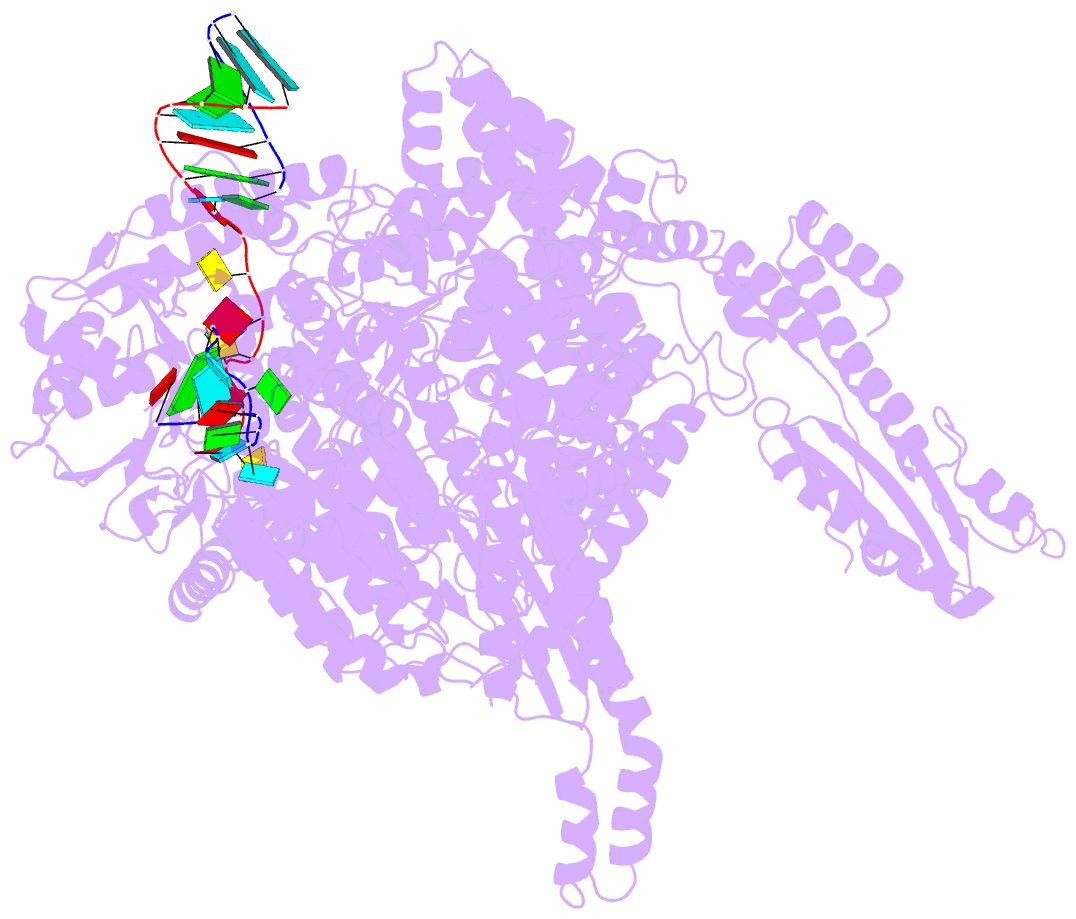
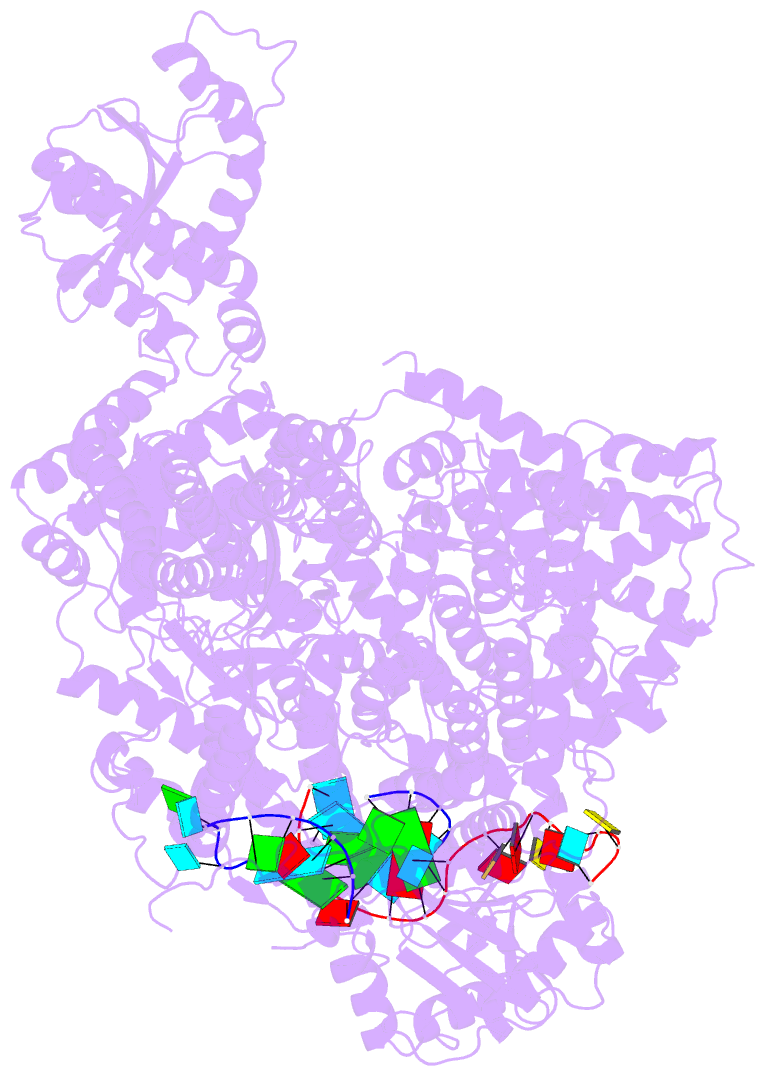
- Structure of the la crosse bunyavirus polymerase in complex with the 3' and 5' viral RNA
- Reference
- Gerlach P, Malet H, Cusack S, Reguera J (2015): "Structural Insights Into Bunyavirus Replication and its Regulation by the Vrna Promoter." Cell, 161, 1267-1279. doi: 10.1016/J.CELL.2015.05.006.
- Abstract
- Segmented negative-strand RNA virus (sNSV) polymerases transcribe and replicate the viral RNA (vRNA) within a ribonucleoprotein particle (RNP). We present cryo-EM and X-ray structures of, respectively, apo- and vRNA bound La Crosse orthobunyavirus (LACV) polymerase that give atomic-resolution insight into how such RNPs perform RNA synthesis. The complementary 3' and 5' vRNA extremities are sequence specifically bound in separate sites on the polymerase. The 5' end binds as a stem-loop, allosterically structuring functionally important polymerase active site loops. Identification of distinct template and product exit tunnels allows proposal of a detailed model for template-directed replication with minimal disruption to the circularised RNP. The similar overall architecture and vRNA binding of monomeric LACV to heterotrimeric influenza polymerase, despite high sequence divergence, suggests that all sNSV polymerases have a common evolutionary origin and mechanism of RNA synthesis. These results will aid development of replication inhibitors of diverse, serious human pathogenic viruses.