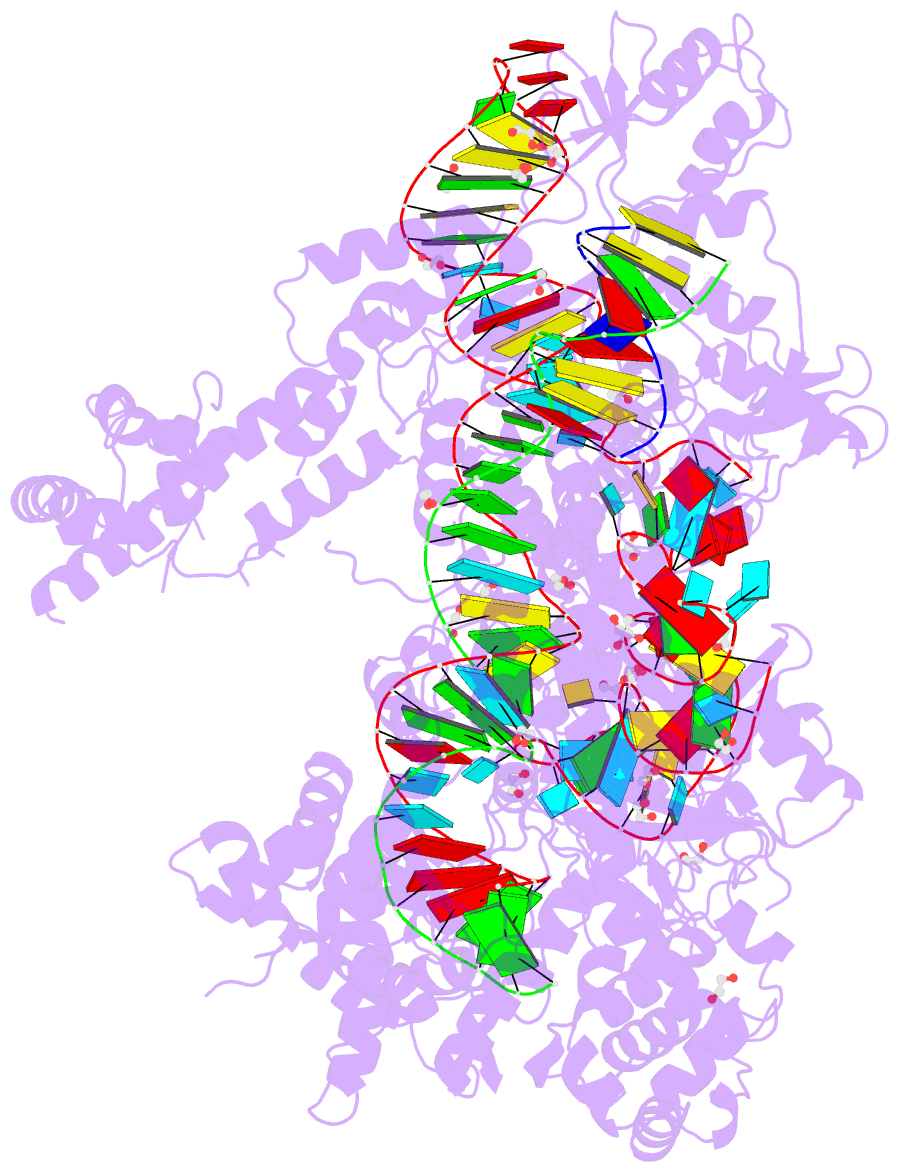
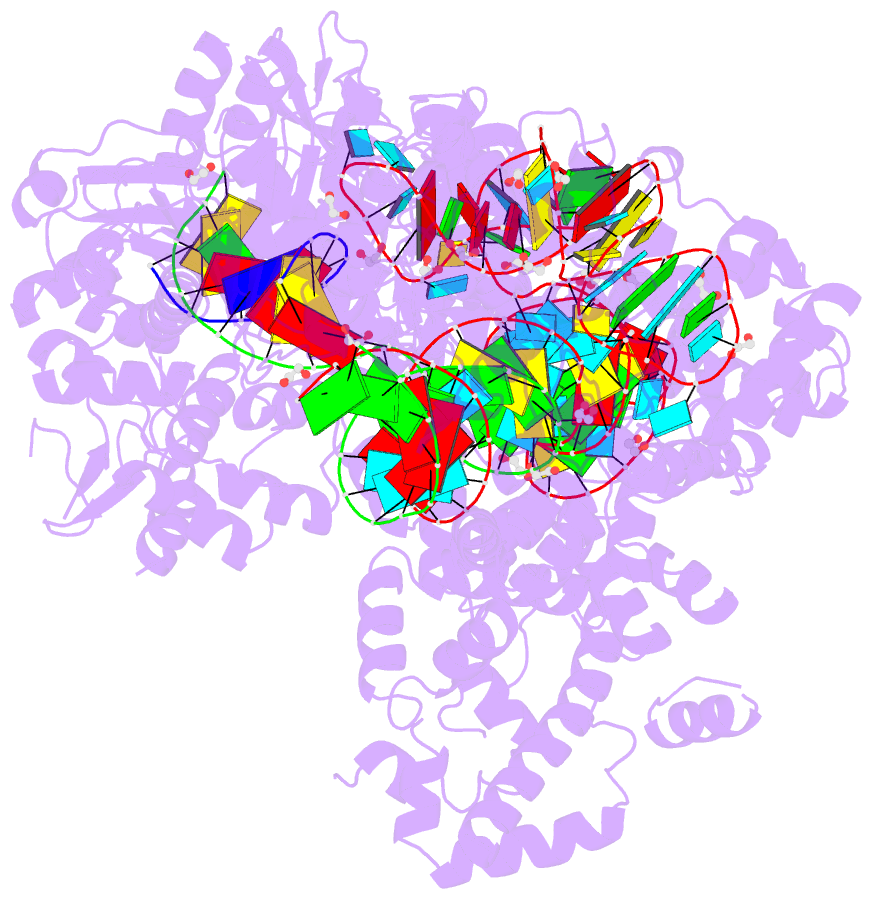
Summary information and primary citation
- PDB-id
- 5b2o; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA-DNA
- Method
- X-ray (1.7 Å)
- Summary
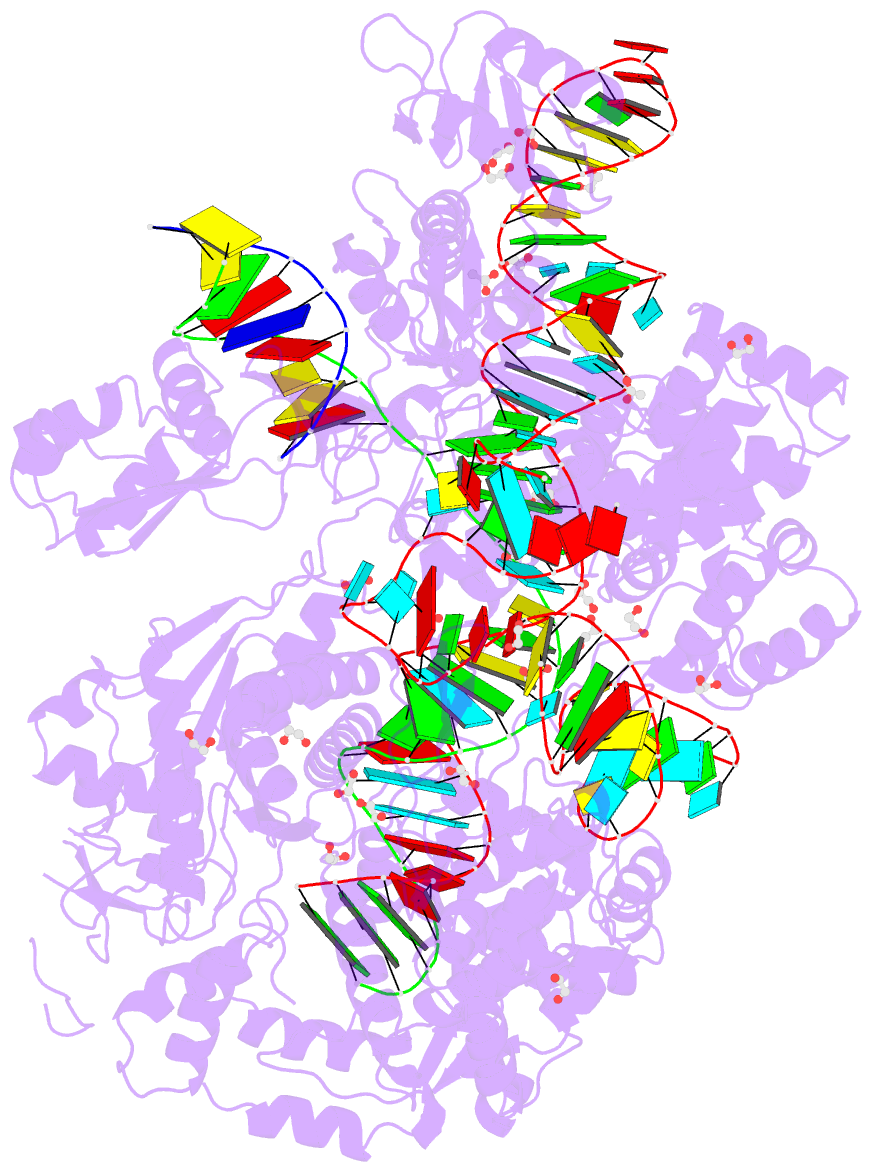
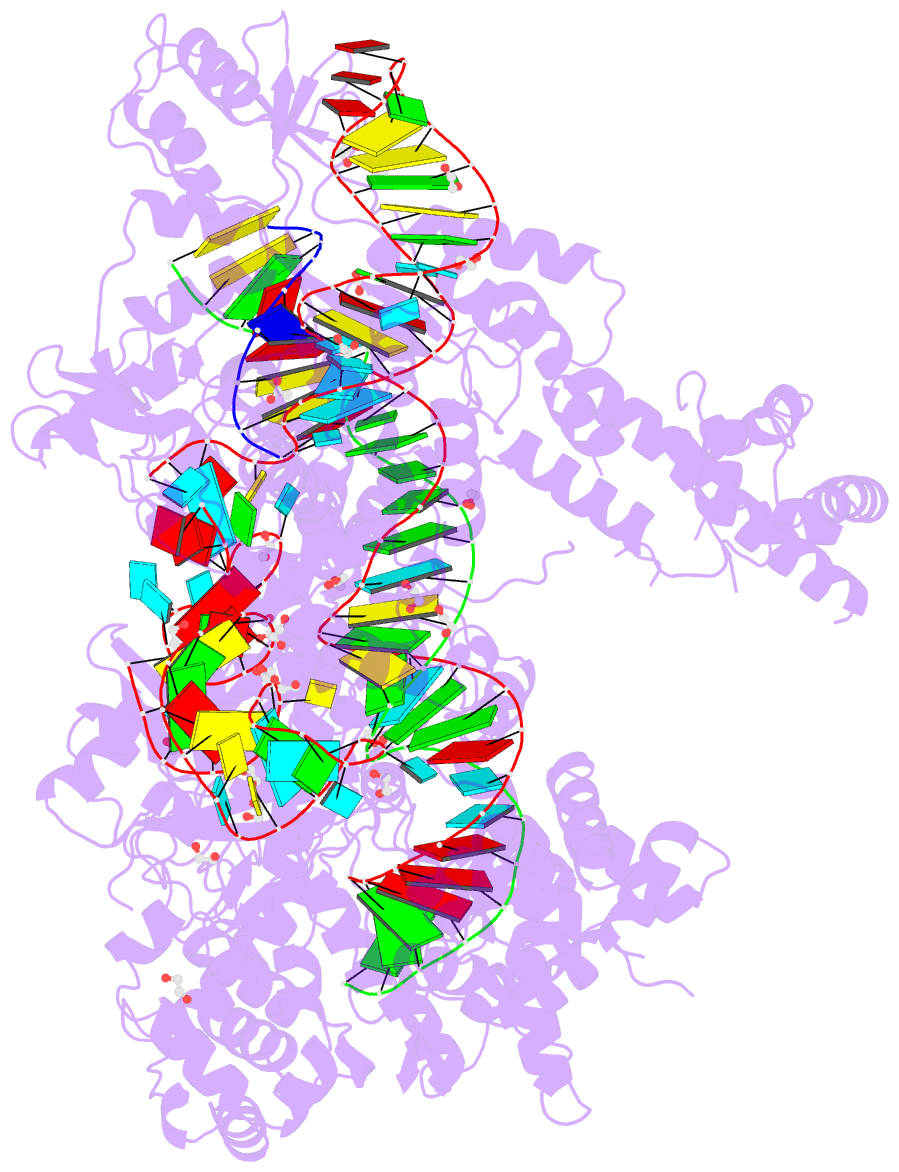
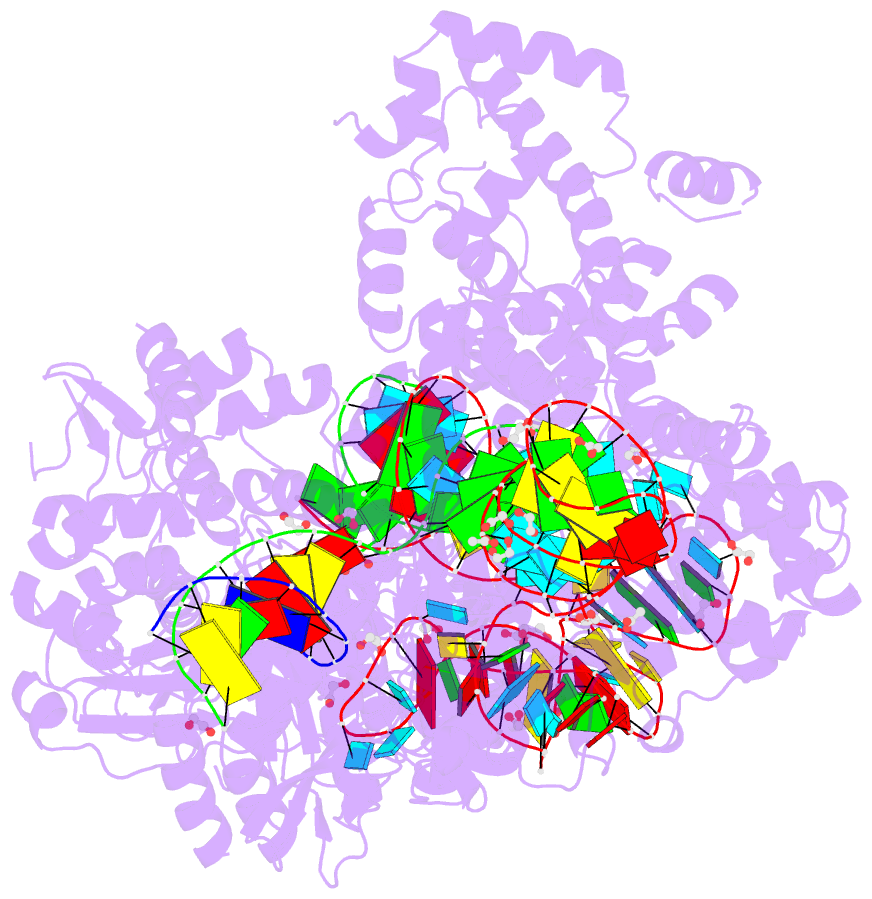
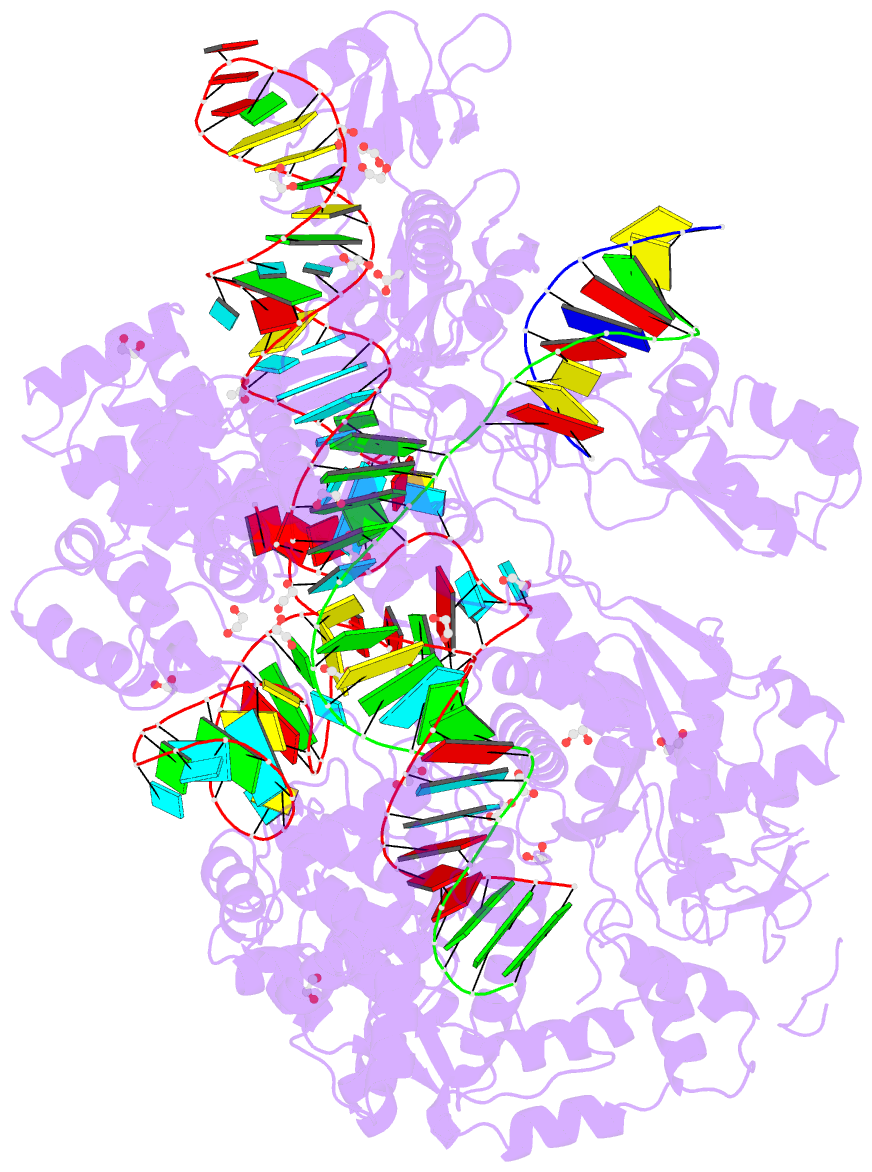
- Crystal structure of francisella novicida cas9 in complex with sgrna and target DNA (tgg pam)
- Reference
- Hirano H, Gootenberg JS, Horii T, Abudayyeh OO, Kimura M, Hsu PD, Nakane T, Ishitani R, Hatada I, Zhang F, Nishimasu H, Nureki O (2016): "Structure and Engineering of Francisella novicida Cas9." Cell, 164, 950-961. doi: 10.1016/j.cell.2016.01.039.
- Abstract
- The RNA-guided endonuclease Cas9 cleaves double-stranded DNA targets complementary to the guide RNA and has been applied to programmable genome editing. Cas9-mediated cleavage requires a protospacer adjacent motif (PAM) juxtaposed with the DNA target sequence, thus constricting the range of targetable sites. Here, we report the 1.7 Å resolution crystal structures of Cas9 from Francisella novicida (FnCas9), one of the largest Cas9 orthologs, in complex with a guide RNA and its PAM-containing DNA targets. A structural comparison of FnCas9 with other Cas9 orthologs revealed striking conserved and divergent features among distantly related CRISPR-Cas9 systems. We found that FnCas9 recognizes the 5'-NGG-3' PAM, and used the structural information to create a variant that can recognize the more relaxed 5'-YG-3' PAM. Furthermore, we demonstrated that the FnCas9-ribonucleoprotein complex can be microinjected into mouse zygotes to edit endogenous sites with the 5'-YG-3' PAM, thus expanding the target space of the CRISPR-Cas9 toolbox.