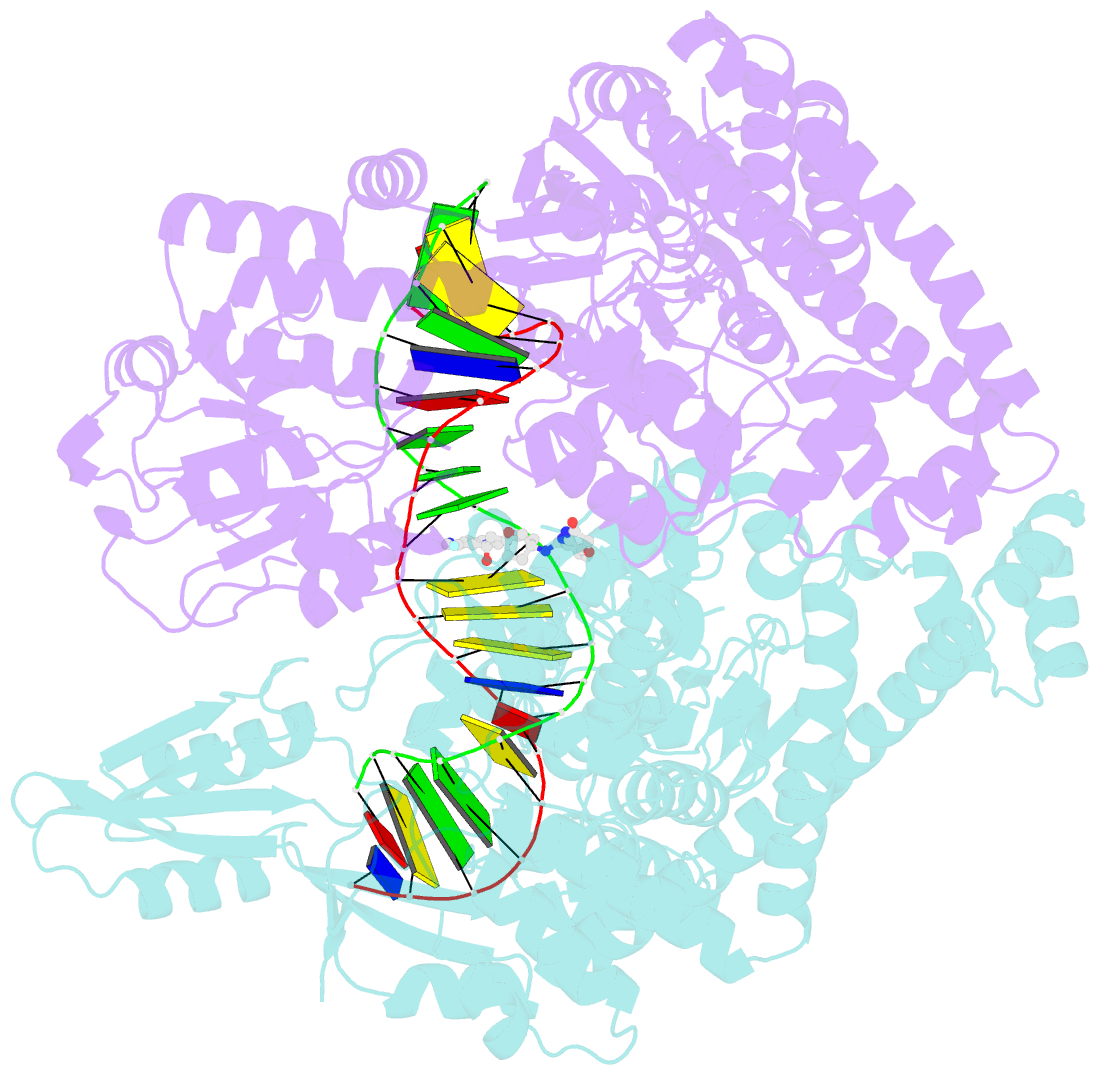
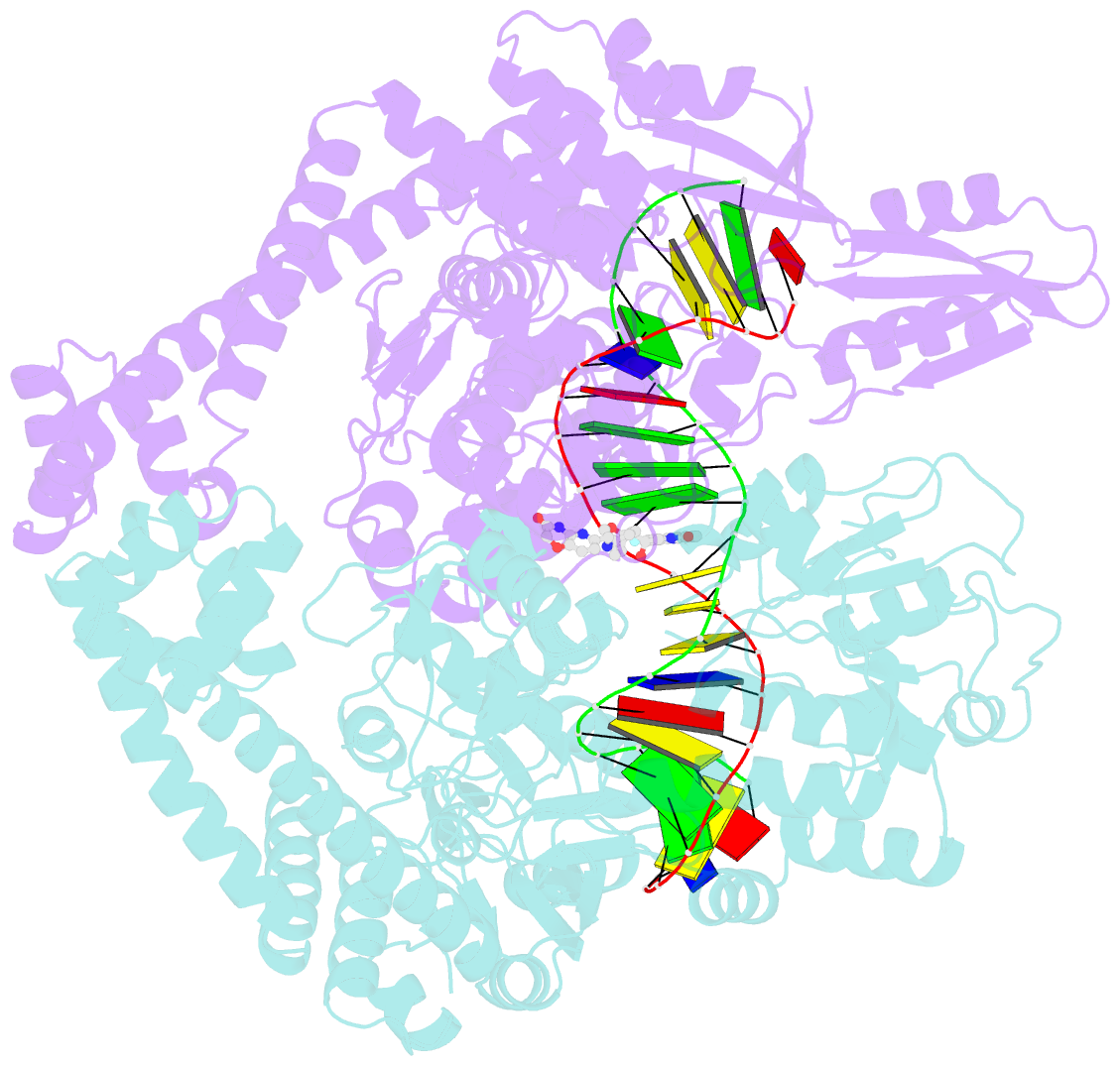
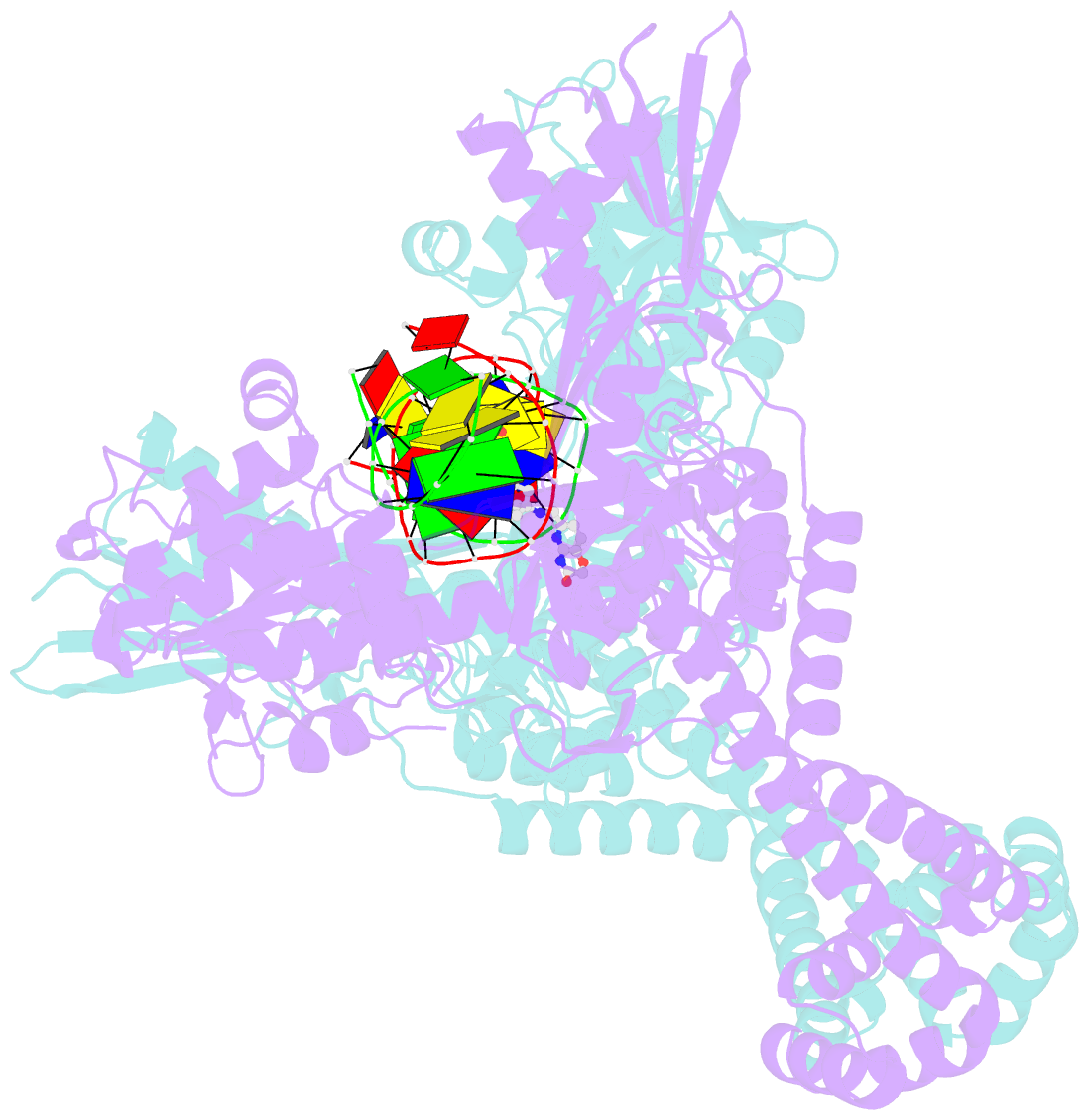
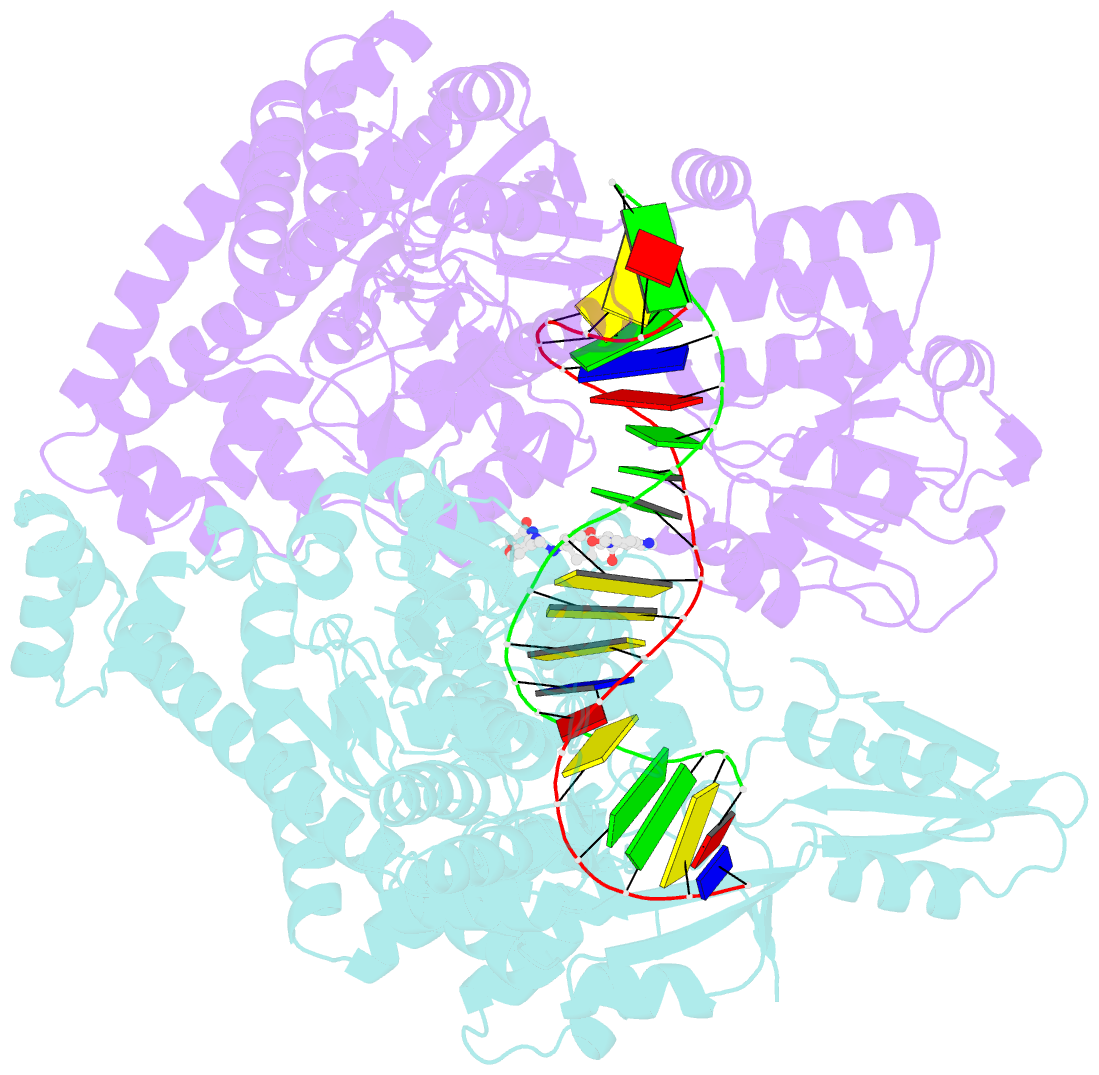
Summary information and primary citation
- PDB-id
- 5bs3; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- isomerase-DNA-RNA
- Method
- X-ray (2.65 Å)
- Summary
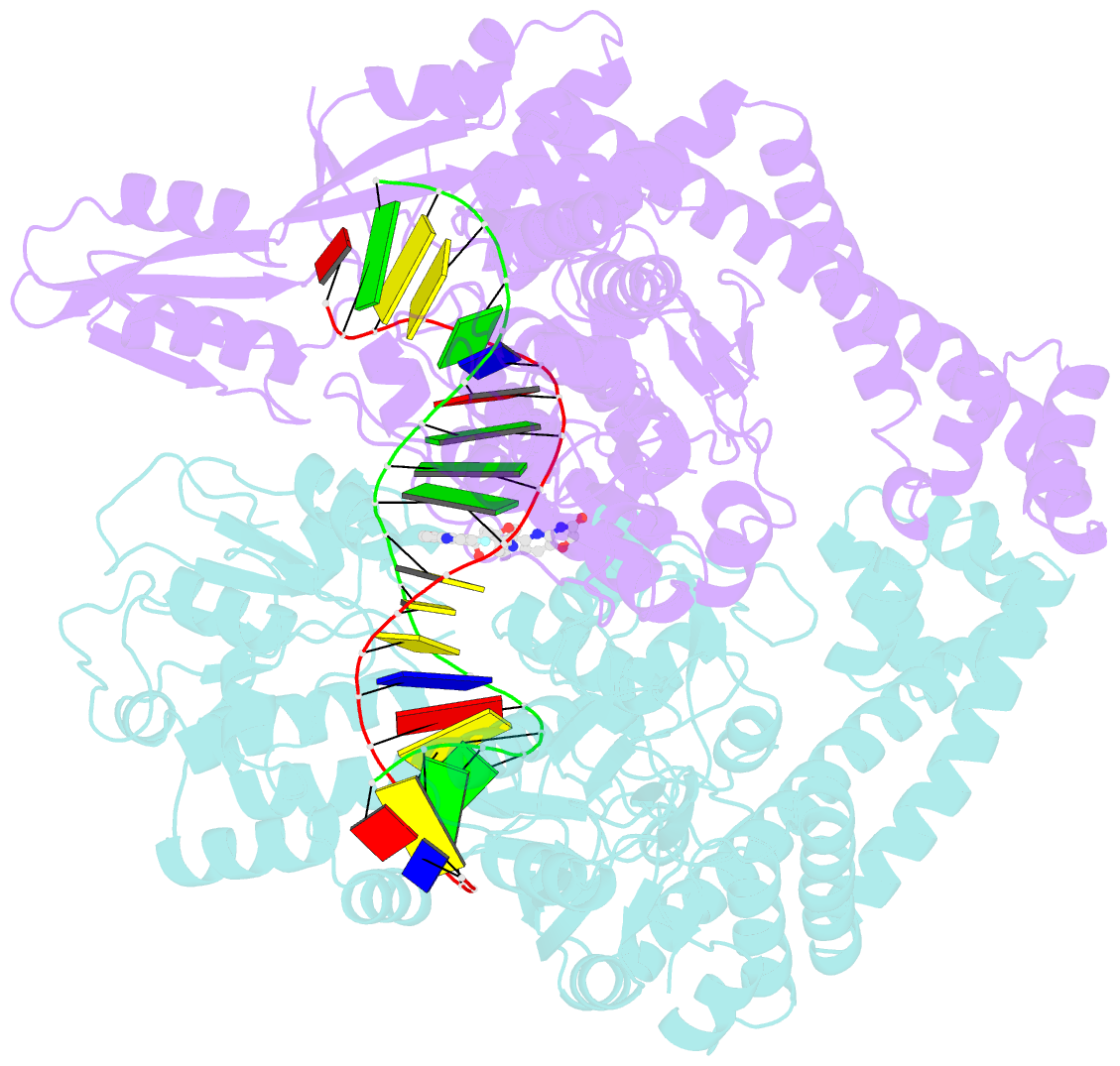
- Crystal structure of s.a. gyrase in complex with compound 7
- Reference
- Singh SB, Kaelin DE, Wu J, Miesel L, Tan CM, Black T, Nargund R, Meinke PT, Olsen DB, Lagrutta A, Lu J, Patel S, Rickert KW, Smith RF, Soisson S, Sherer E, Joyce LA, Wei C, Peng X, Wang X, Fukuda H, Kishii R, Takei M, Takano H, Shibasaki M, Yajima M, Nishimura A, Shibata T, Fukuda Y (2015): "Tricyclic 1,5-naphthyridinone oxabicyclooctane-linked novel bacterial topoisomerase inhibitors as broad-spectrum antibacterial agents-SAR of left-hand-side moiety (Part-2)." Bioorg.Med.Chem.Lett., 25, 1831-1835. doi: 10.1016/j.bmcl.2015.03.044.
- Abstract
- Novel bacterial topoisomerase inhibitors (NBTIs) represent a new class of broad-spectrum antibacterial agents targeting bacterial Gyrase A and ParC and have potential utility in combating antibiotic resistance. A series of novel oxabicyclooctane-linked NBTIs with new tricyclic-1,5-naphthyridinone left hand side moieties have been described. Compounds with a (R)-hydroxy-1,5-naphthyridinone moiety (7) showed potent antibacterial activity (e.g., Staphylococcus aureus MIC 0.25 μg/mL), acceptable Gram-positive and Gram-negative spectrum with rapidly bactericidal activity. The compound 7 showed intravenous and oral efficacy (ED50) at 3.2 and 27 mg/kg doses, respectively, in a murine model of bacteremia. Most importantly they showed significant attenuation of functional hERG activity (IC50 >170 μM). In general, lower logD attenuated hERG activity but also reduced Gram-negative activity. The co-crystal structure of a hydroxy-tricyclic NBTI bound to a DNA-gyrase complex exhibited a binding mode that show enantiomeric preference for R isomer and explains the activity and SAR. The discovery, synthesis, SAR and X-ray crystal structure of the left-hand-side tricyclic 1,5-naphthyridinone based oxabicyclooctane linked NBTIs are described.