Summary information and primary citation
- PDB-id
- 5bt2; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- DNA binding protein-DNA
- Method
- X-ray (2.2 Å)
- Summary
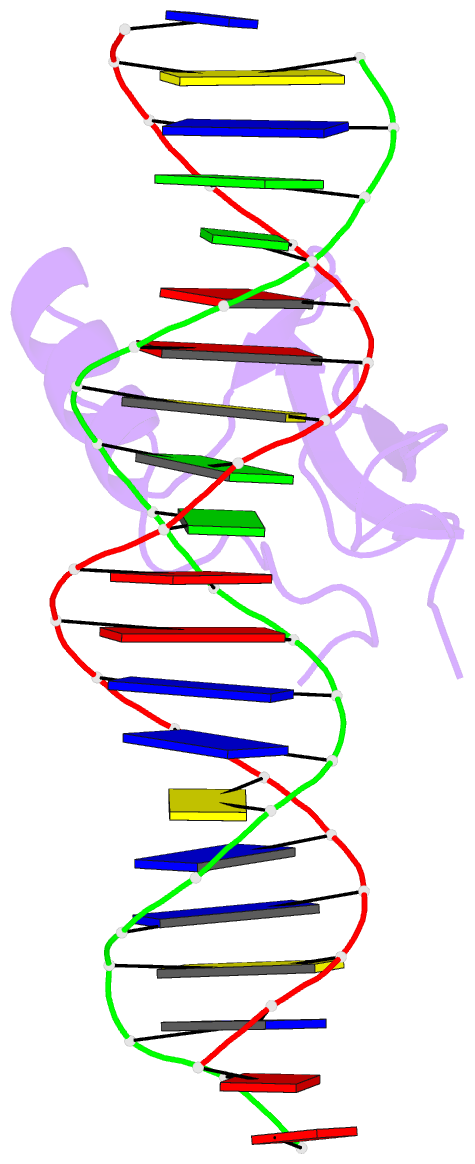
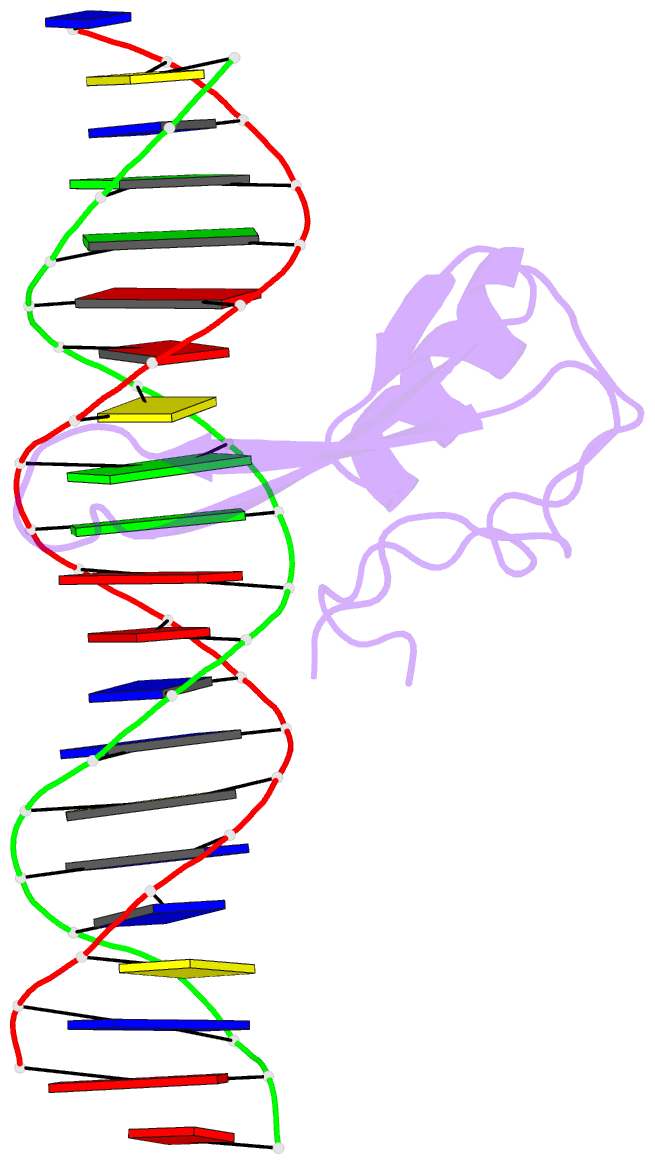
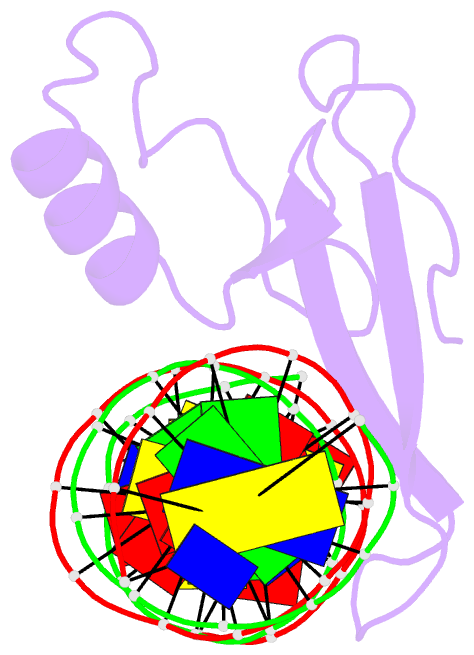
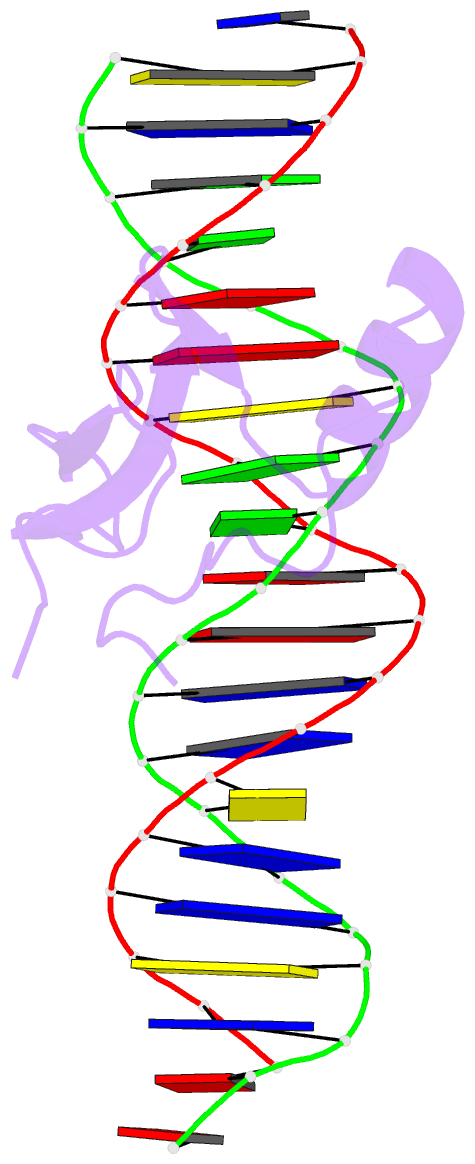
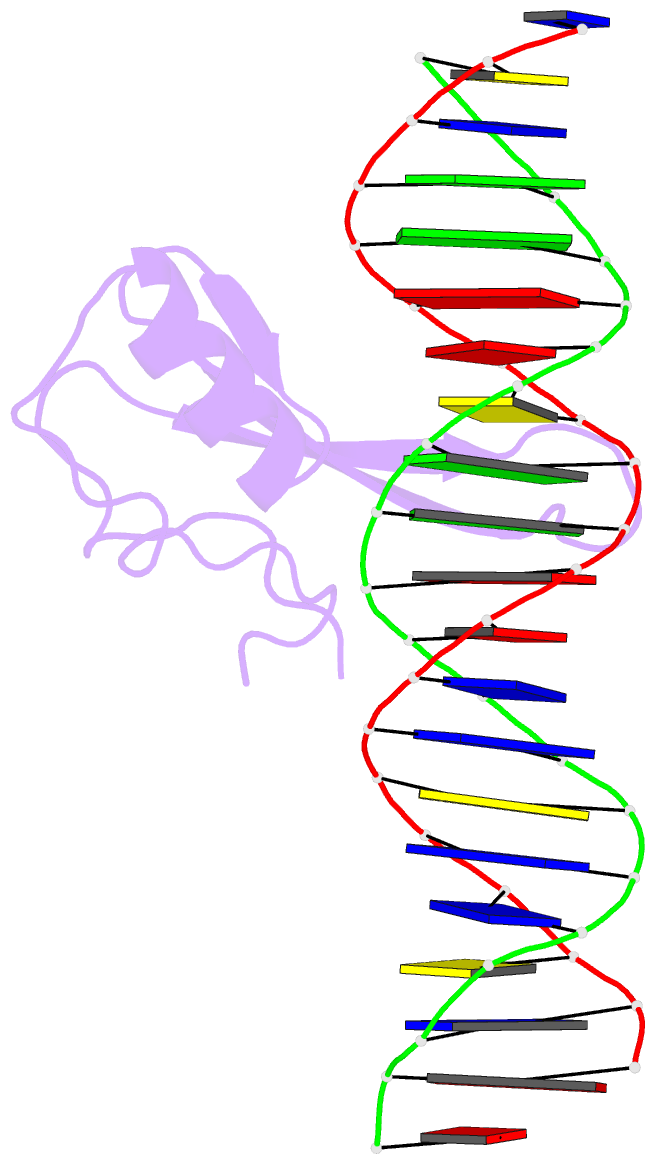
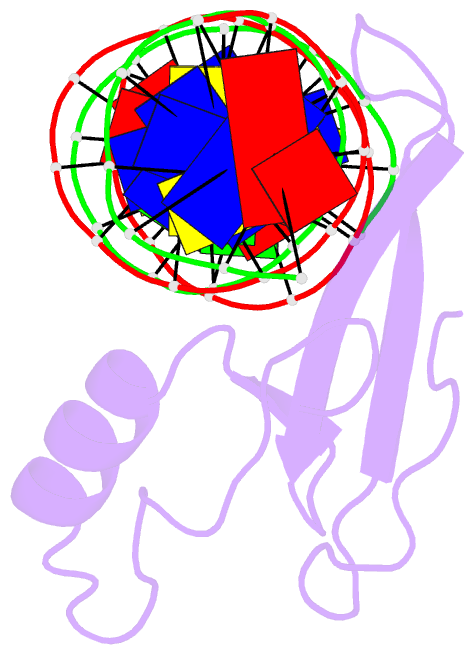
- Mecp2 mbd domain (a140v) in complex with methylated DNA
- Reference
- Chia JY, Tan WS, Ng CL, Hu NJ, Foo HL, Ho KL (2016): "A/T Run Geometry of B-form DNA Is Independent of Bound Methyl-CpG Binding Domain, Cytosine Methylation and Flanking Sequence." Sci Rep, 6, 31210. doi: 10.1038/srep31210.
- Abstract
- DNA methylation in a CpG context can be recognised by methyl-CpG binding protein 2 (MeCP2) via its methyl-CpG binding domain (MBD). An A/T run next to a methyl-CpG maximises the binding of MeCP2 to the methylated DNA. The A/T run characteristics are reported here with an X-ray structure of MBD A140V in complex with methylated DNA. The A/T run geometry was found to be strongly stabilised by a string of conserved water molecules regardless of its flanking nucleotide sequences, DNA methylation and bound MBD. New water molecules were found to stabilise the Rett syndrome-related E137, whose carboxylate group is salt bridged to R133. A structural comparison showed no difference between the wild type and MBD A140V. However, differential scanning calorimetry showed that the melting temperature of A140V constructs in complex with methylated DNA was reduced by ~7 °C, although circular dichroism showed no changes in the secondary structure content for A140V. A band shift analysis demonstrated that the larger fragment of MeCP2 (A140V) containing the transcriptional repression domain (TRD) destabilises the DNA binding. These results suggest that the solution structure of MBD A140V may differ from the wild-type MBD although no changes in the biochemical properties of X-ray A140V were observed.