Summary information and primary citation
- PDB-id
- 5bte; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- hydrolase-RNA
- Method
- X-ray (2.4 Å)
- Summary
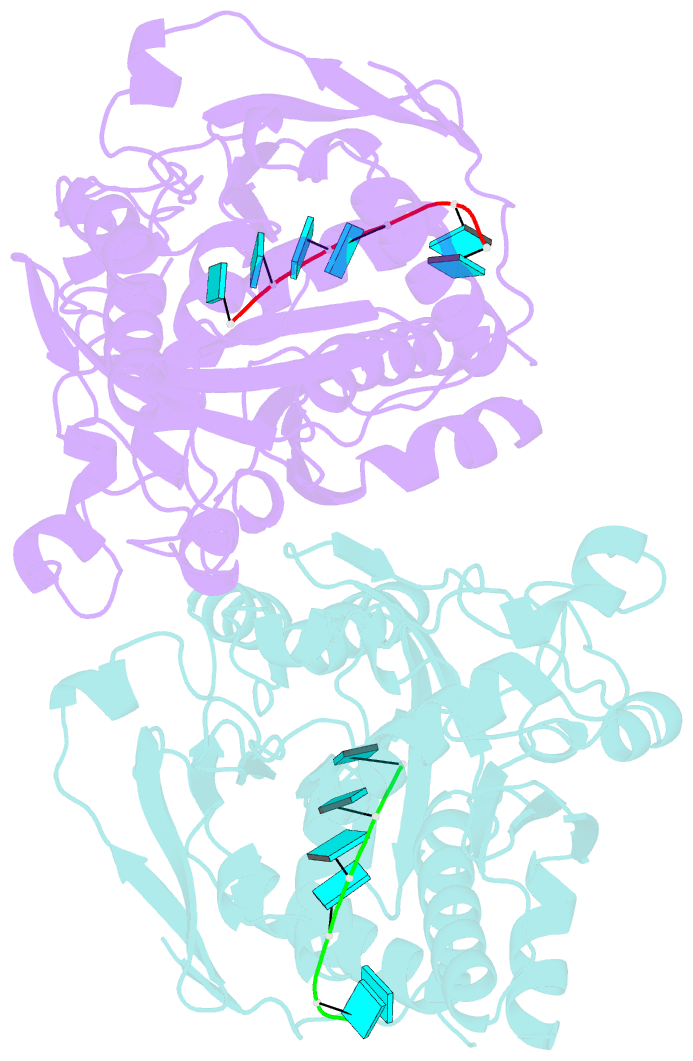
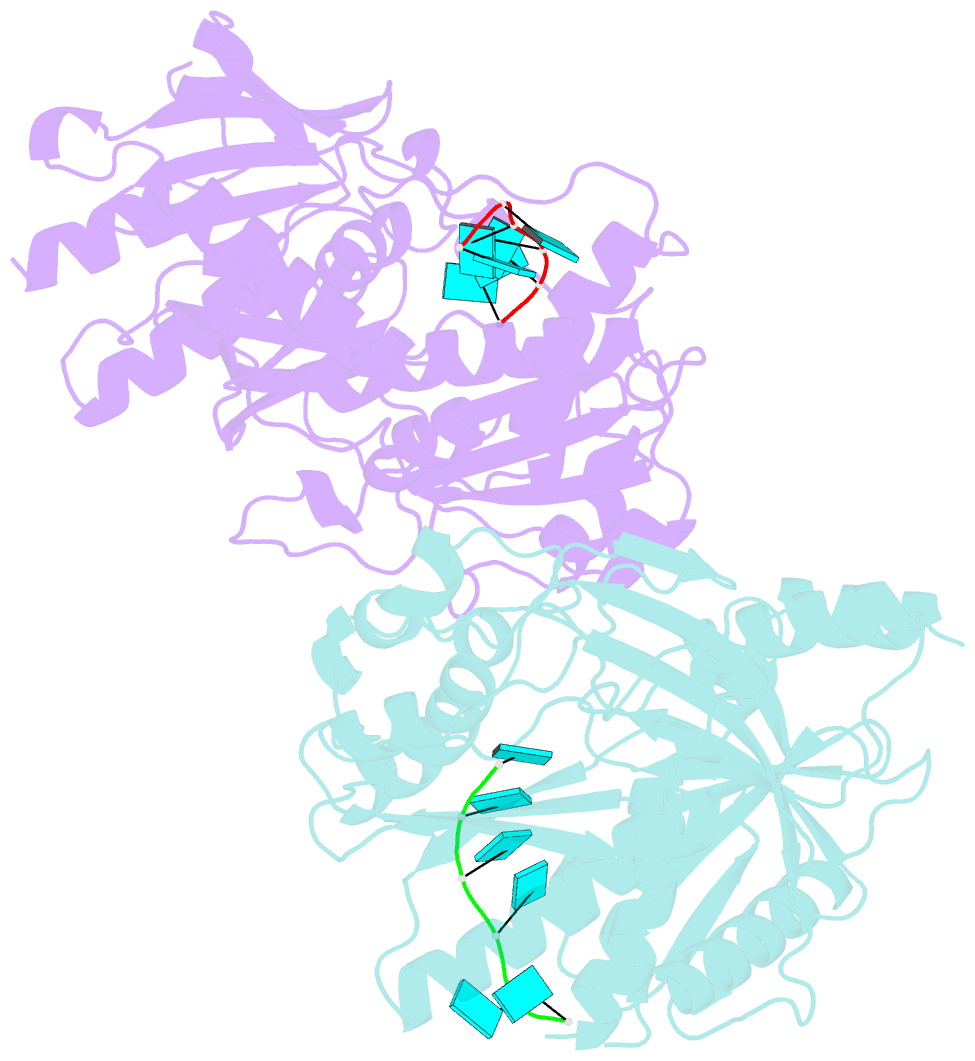
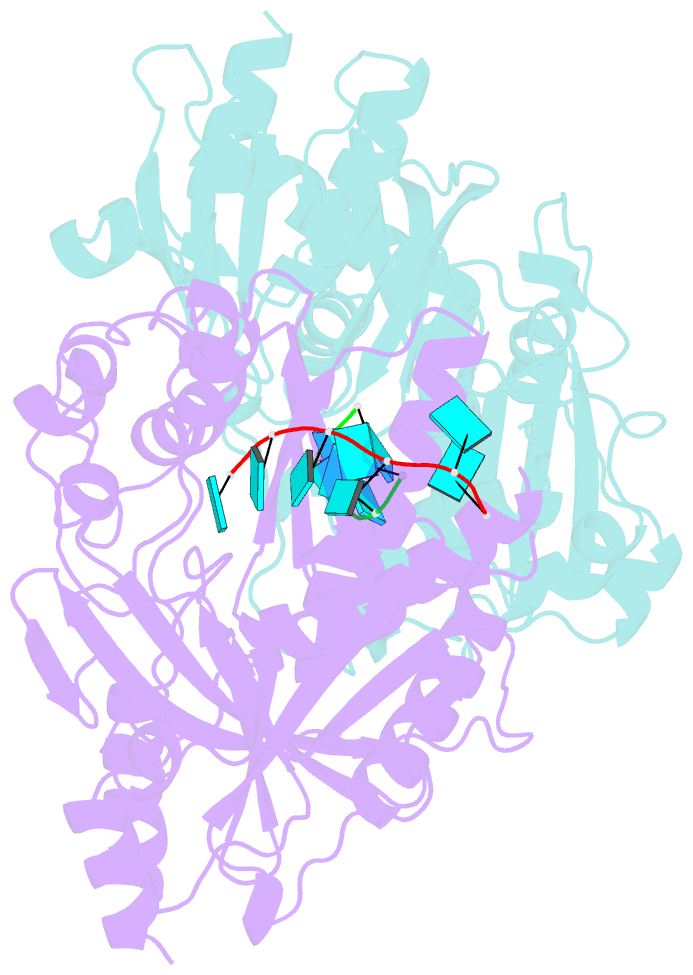
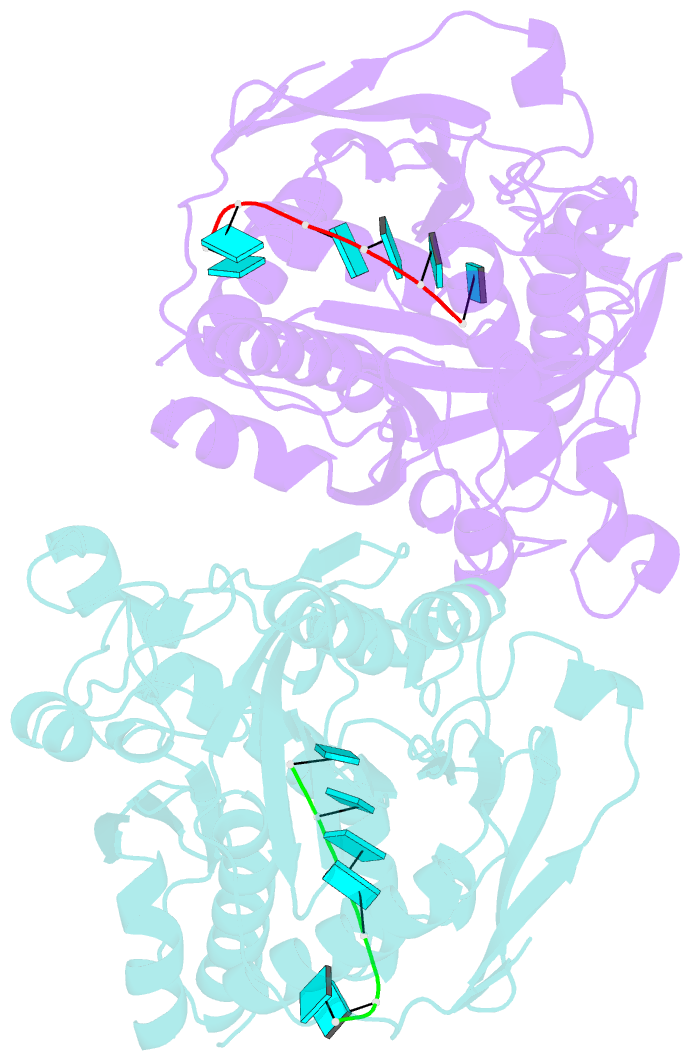
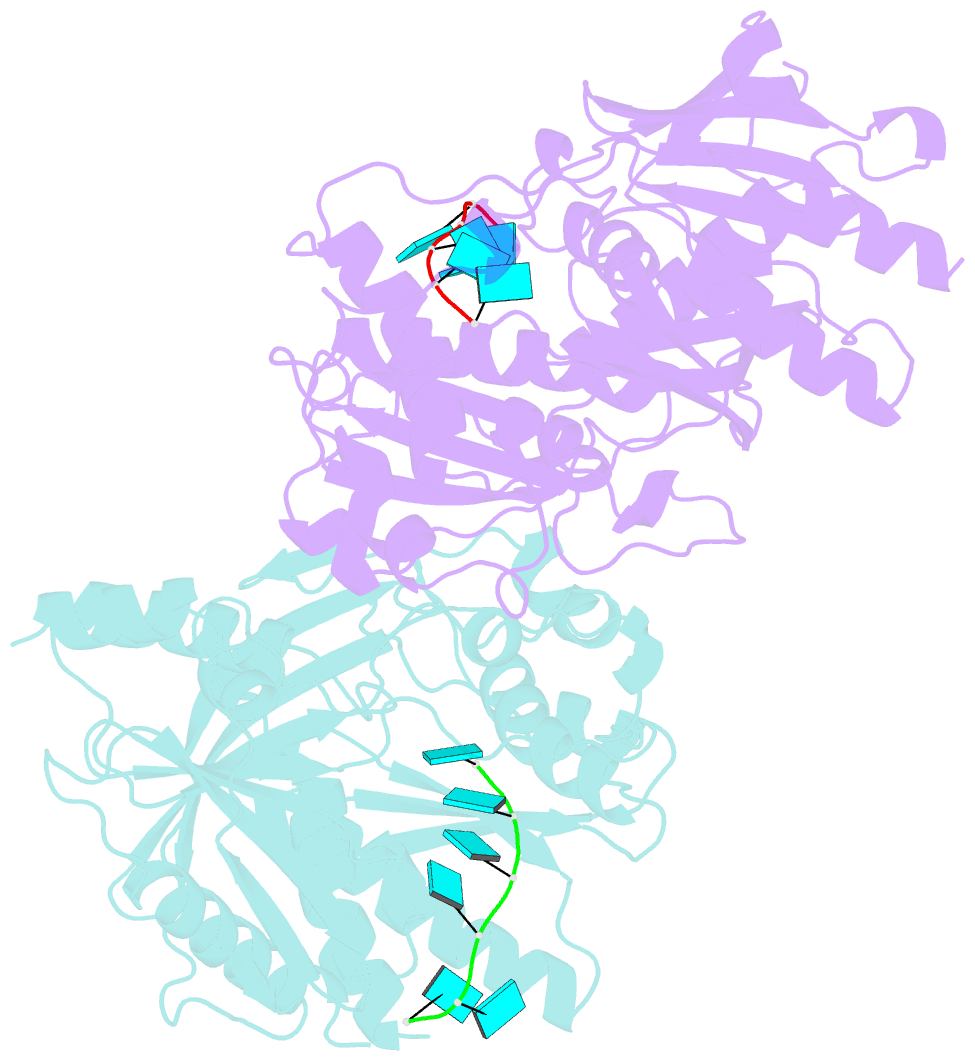
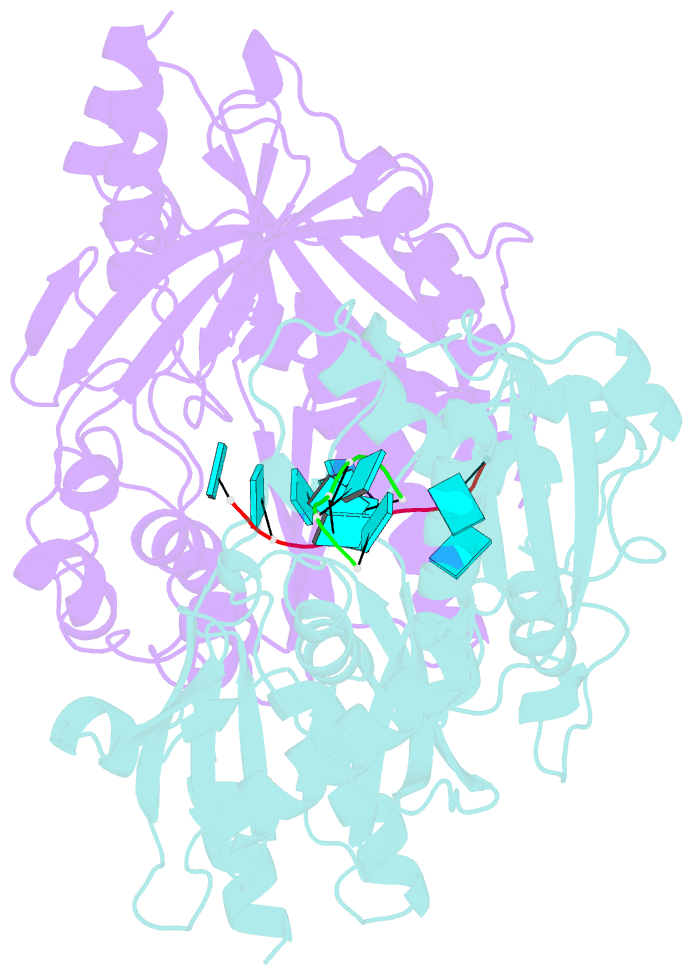
- Crystal structure of ashbya gossypii rai1 in complex with pu(s)6-mn2+
- Reference
- Wang VY, Jiao X, Kiledjian M, Tong L (2015): "Structural and biochemical studies of the distinct activity profiles of Rai1 enzymes." Nucleic Acids Res., 43, 6596-6606. doi: 10.1093/nar/gkv620.
- Abstract
- Recent studies showed that Rai1 and its homologs are a crucial component of the mRNA 5'-end capping quality control mechanism. They can possess RNA 5'-end pyrophosphohydrolase (PPH), decapping, and 5'-3' exonuclease (toward 5' monophosphate RNA) activities, which help to degrade mRNAs with incomplete 5'-end capping. A single active site in the enzyme supports these apparently distinct activities. However, each Rai1 protein studied so far has a unique set of activities, and the molecular basis for these differences are not known. Here, we have characterized the highly diverse activity profiles of Rai1 homologs from a collection of fungal organisms and identified a new activity for these enzymes, 5'-end triphosphonucleotide hydrolase (TPH) instead of PPH activity. Crystal structures of two of these enzymes bound to RNA oligonucleotides reveal differences in the RNA binding modes. Structure-based mutations of these enzymes, changing residues that contact the RNA but are poorly conserved, have substantial effects on their activity, providing a framework to begin to understand the molecular basis for the different activity profiles.