Summary information and primary citation
- PDB-id
- 5bua; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription
- Method
- X-ray (1.812 Å)
- Summary
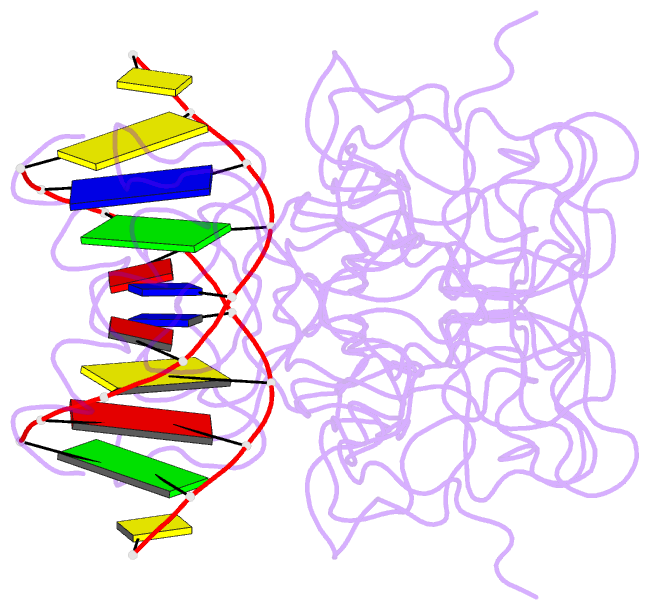
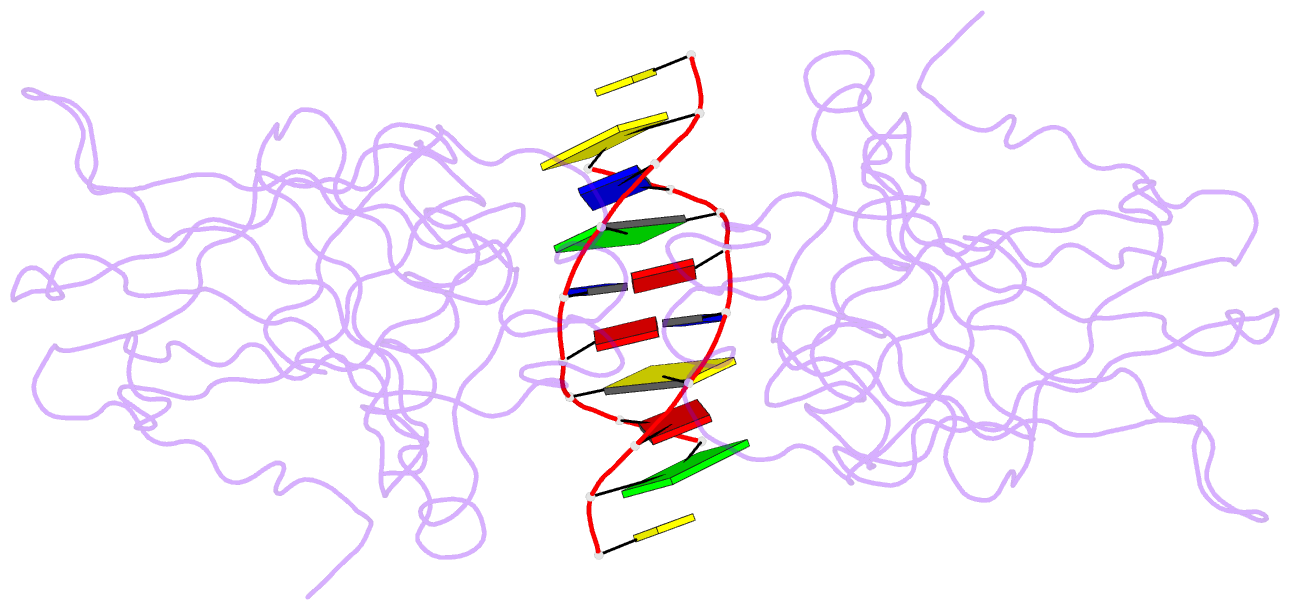
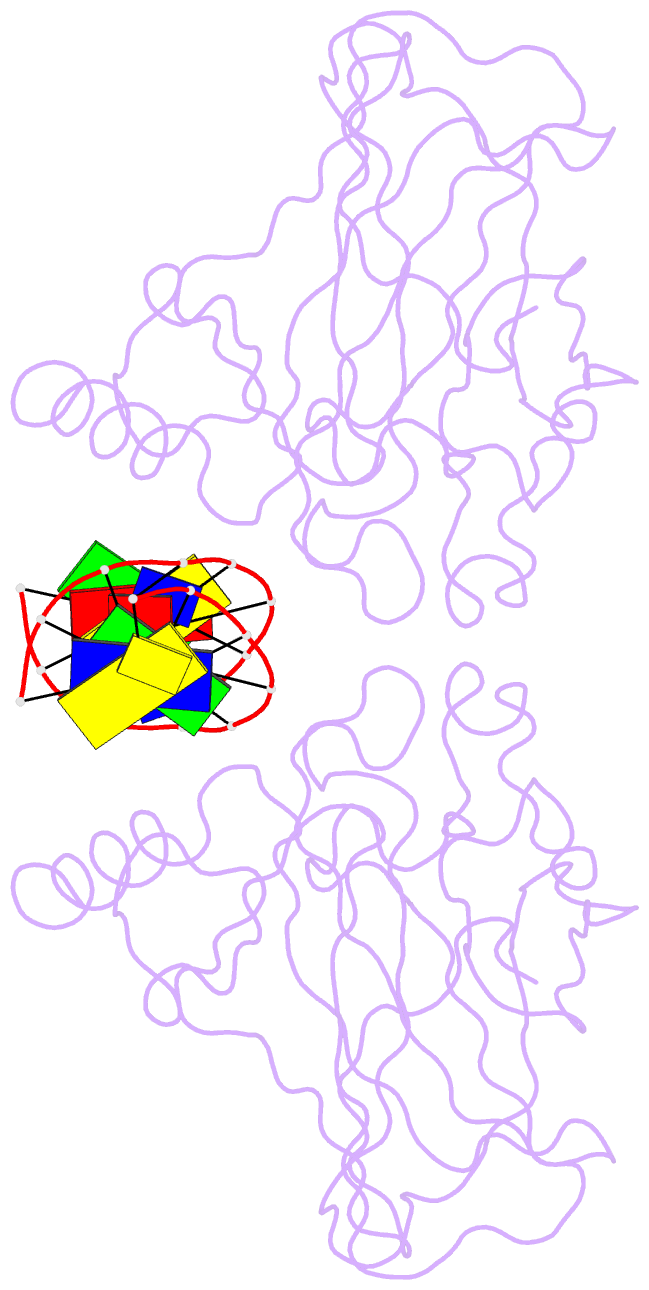
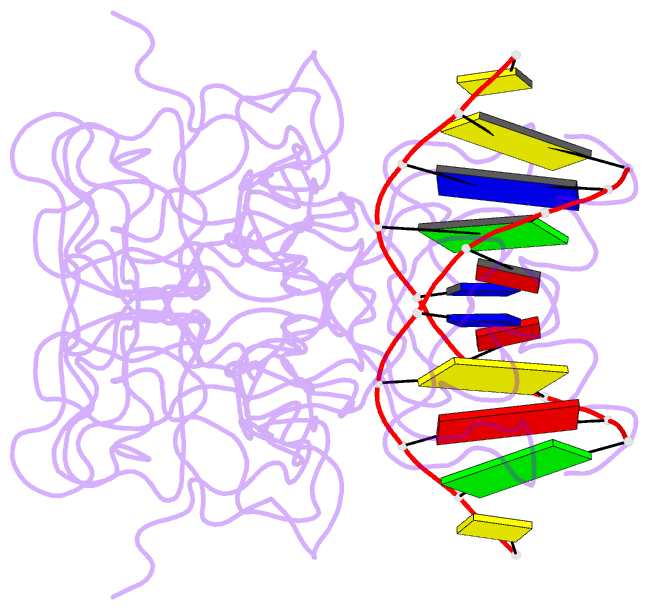
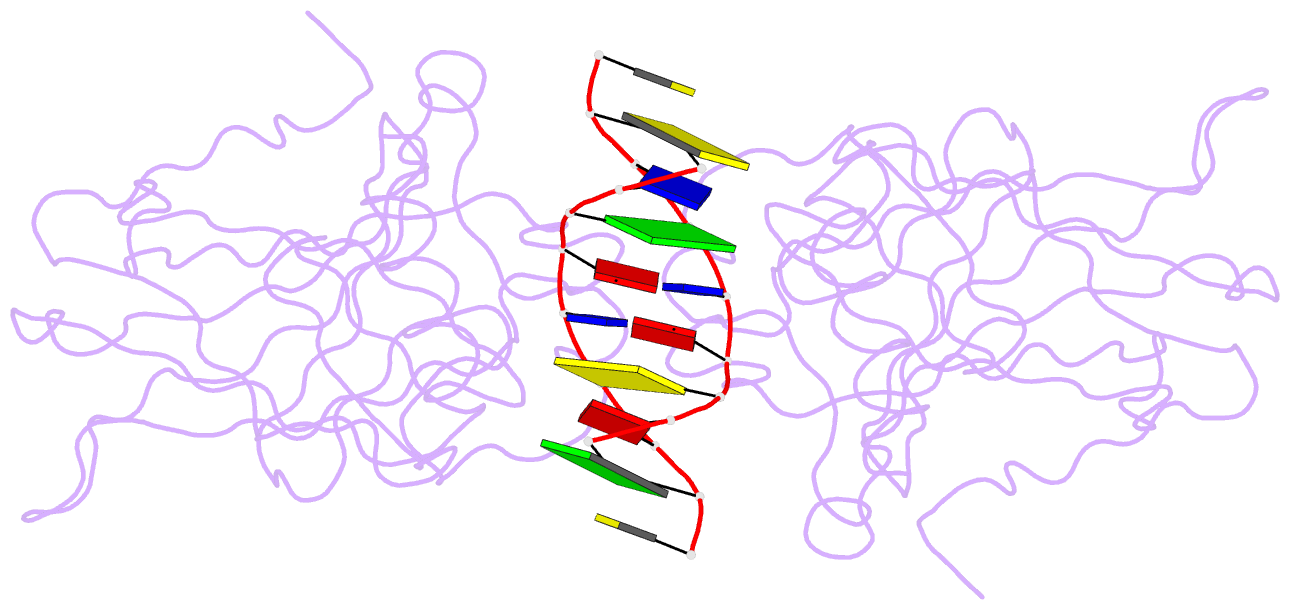
- Lysine 120-acetylated p53 DNA binding domain in a complex with DNA.
- Reference
- Vainer R, Cohen S, Shahar A, Zarivach R, Arbely E (2016): "Structural Basis for p53 Lys120-Acetylation-Dependent DNA-Binding Mode." J.Mol.Biol., 428, 3013-3025. doi: 10.1016/j.jmb.2016.06.009.
- Abstract
- Normal cellular homeostasis depends on tight regulation of gene expression, which requires the modulation of transcription factors' DNA-binding specificity. That said, the mechanisms that allow transcription factors to distinguish between closely related response elements following different cellular signals are not fully understood. In the tumor suppressor protein p53, acetylation of loop L1 residue Lys120 within the DNA-binding domain has been shown to promote the transcription of proapoptotic genes such as bax. Here, we report the crystal structures of Lys120-acetylated p53 DNA-binding domain in complex with a consensus response element and with the natural BAX response element. Our structural analyses reveal that Lys120 acetylation expands the conformational space of loop L1 in the DNA-bound state. Loop L1 flexibility is known to increase p53's DNA-binding specificity, and Lys120-acetylation-dependent conformational changes in loop L1 enable the formation of sequence-dependent DNA-binding modes for p53. Furthermore, binding to the natural BAX response element is accompanied by global conformational changes, deformation of the DNA helical structure, and formation of an asymmetric tetrameric complex. Based on these findings, we suggest a model for p53's Lys120 acetylation-dependent DNA-binding mode.