Summary information and primary citation
- PDB-id
- 5bz5; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (2.8 Å)
- Summary
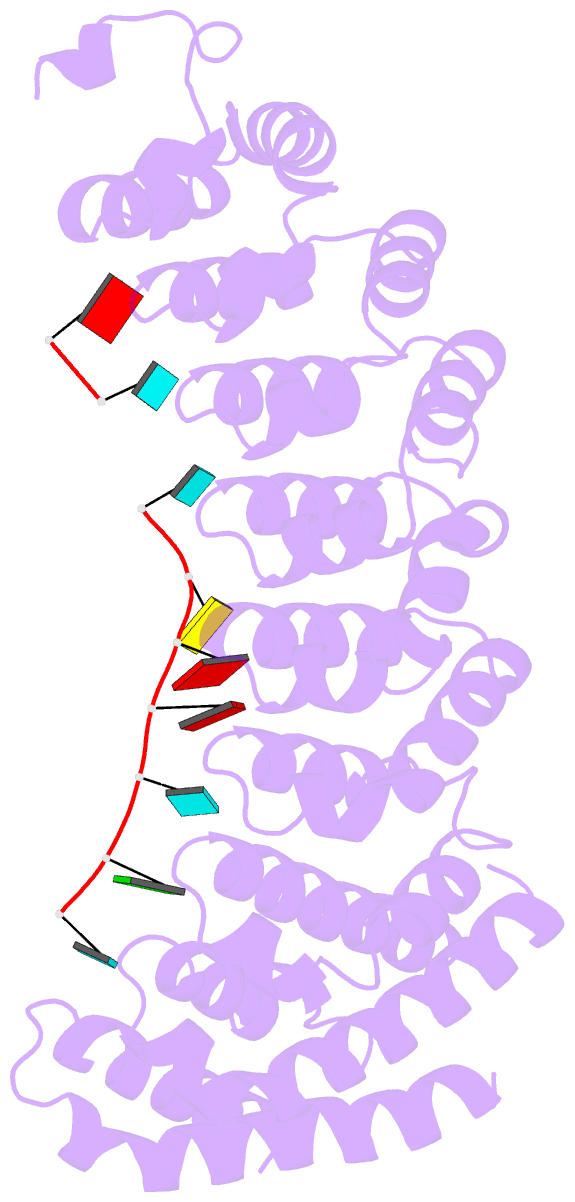
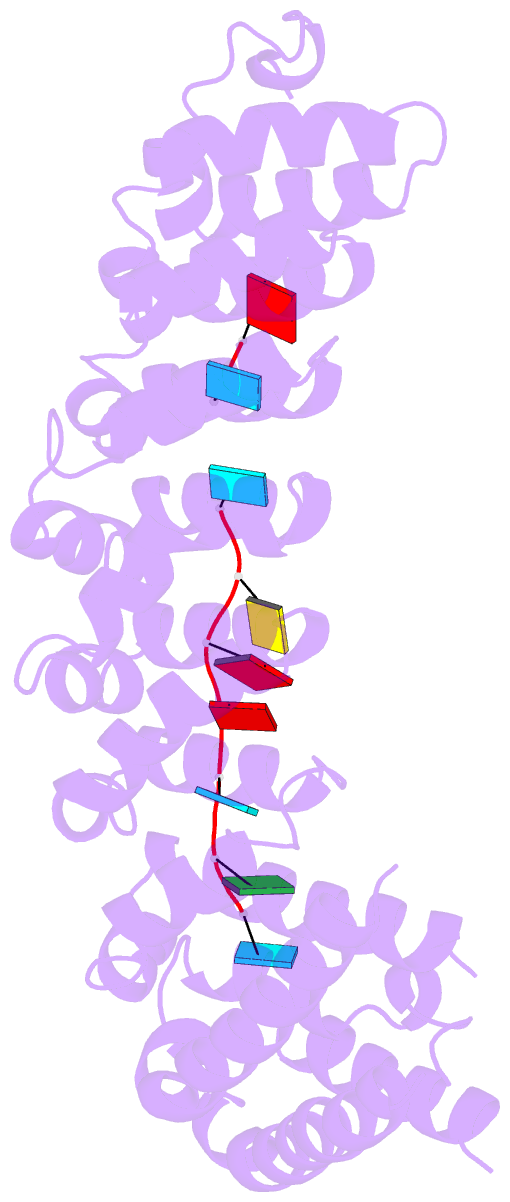
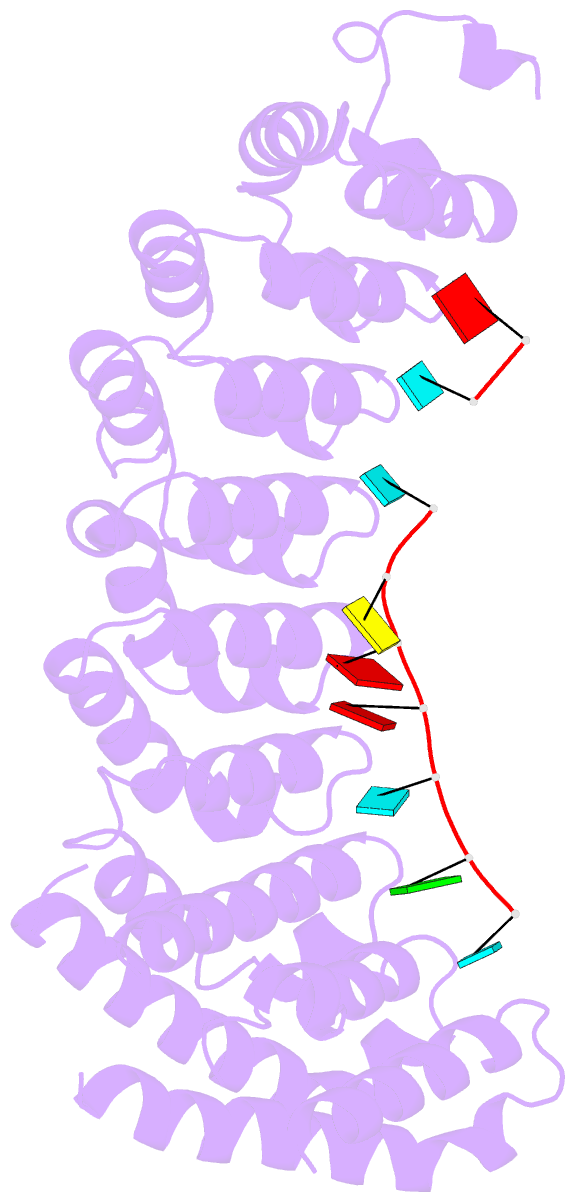
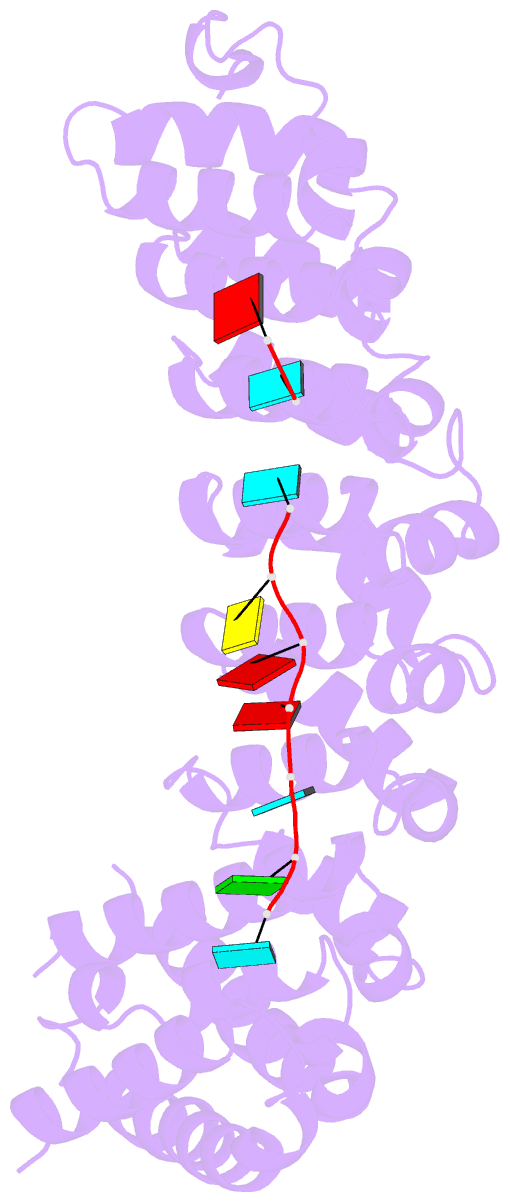
- Crystal structure of the RNA-binding domain of yeast puf5p bound to amn1 RNA
- Reference
- Wilinski D, Qiu C, Lapointe CP, Nevil M, Campbell ZT, Tanaka Hall TM, Wickens M (2015): "RNA regulatory networks diversified through curvature of the PUF protein scaffold." Nat Commun, 6, 8213. doi: 10.1038/ncomms9213.
- Abstract
- Proteins bind and control mRNAs, directing their localization, translation and stability. Members of the PUF family of RNA-binding proteins control multiple mRNAs in a single cell, and play key roles in development, stem cell maintenance and memory formation. Here we identified the mRNA targets of a S. cerevisiae PUF protein, Puf5p, by ultraviolet-crosslinking-affinity purification and high-throughput sequencing (HITS-CLIP). The binding sites recognized by Puf5p are diverse, with variable spacer lengths between two specific sequences. Each length of site correlates with a distinct biological function. Crystal structures of Puf5p-RNA complexes reveal that the protein scaffold presents an exceptionally flat and extended interaction surface relative to other PUF proteins. In complexes with RNAs of different lengths, the protein is unchanged. A single PUF protein repeat is sufficient to induce broadening of specificity. Changes in protein architecture, such as alterations in curvature, may lead to evolution of mRNA regulatory networks.