Summary information and primary citation
- PDB-id
- 5c44; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA-RNA
- Method
- X-ray (3.95 Å)
- Summary
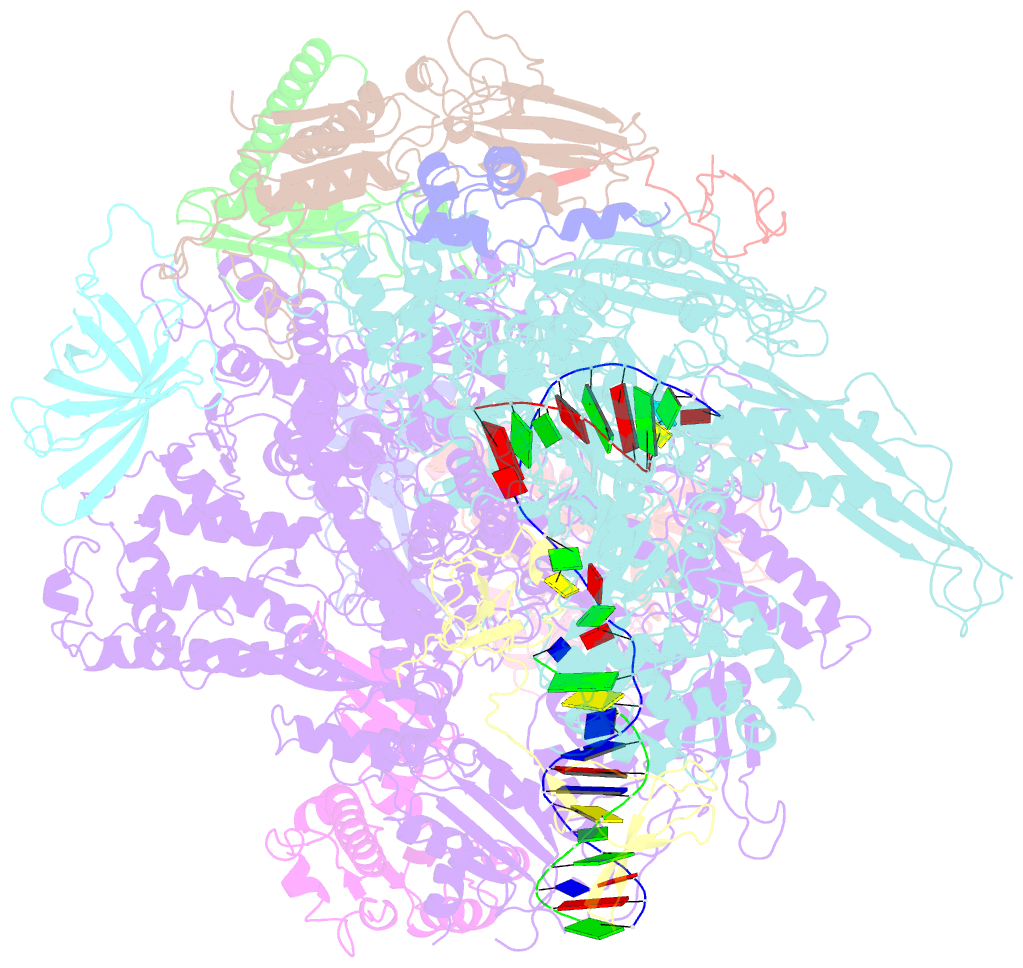
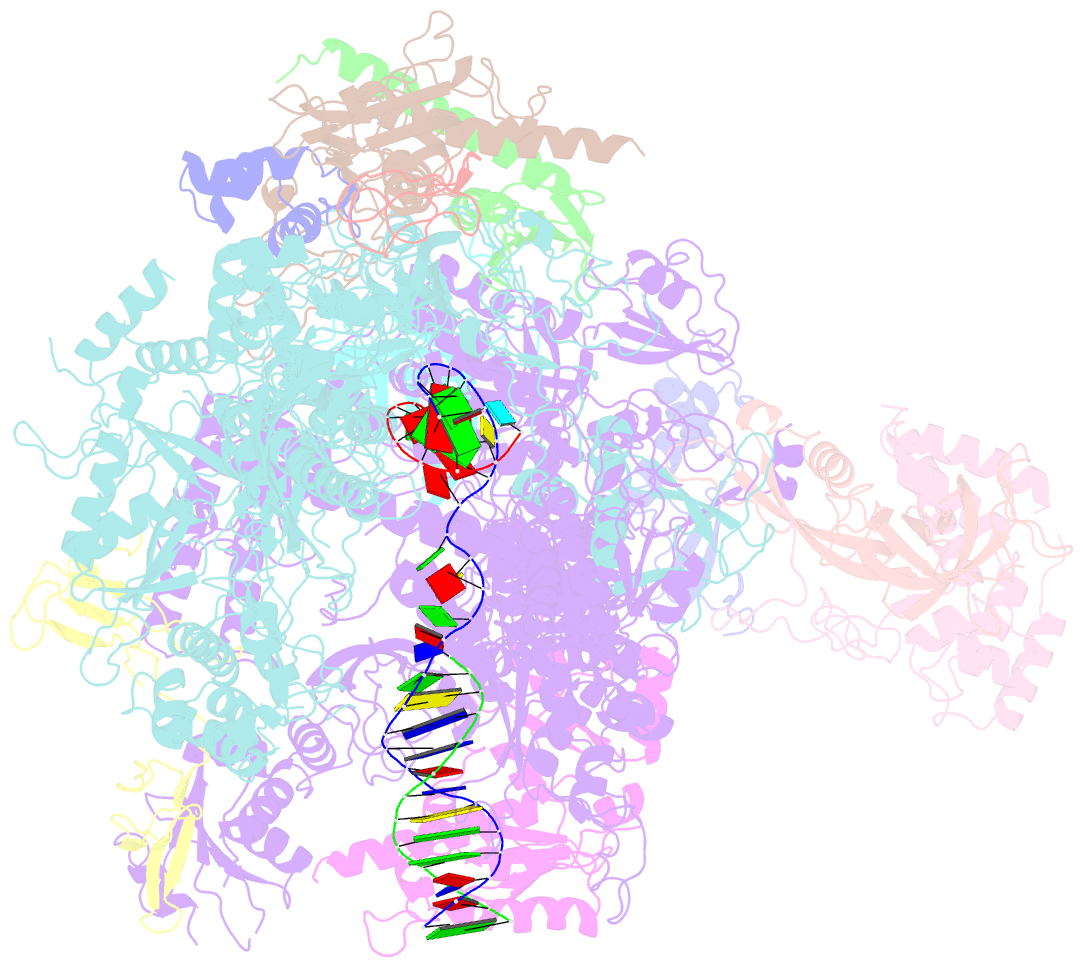
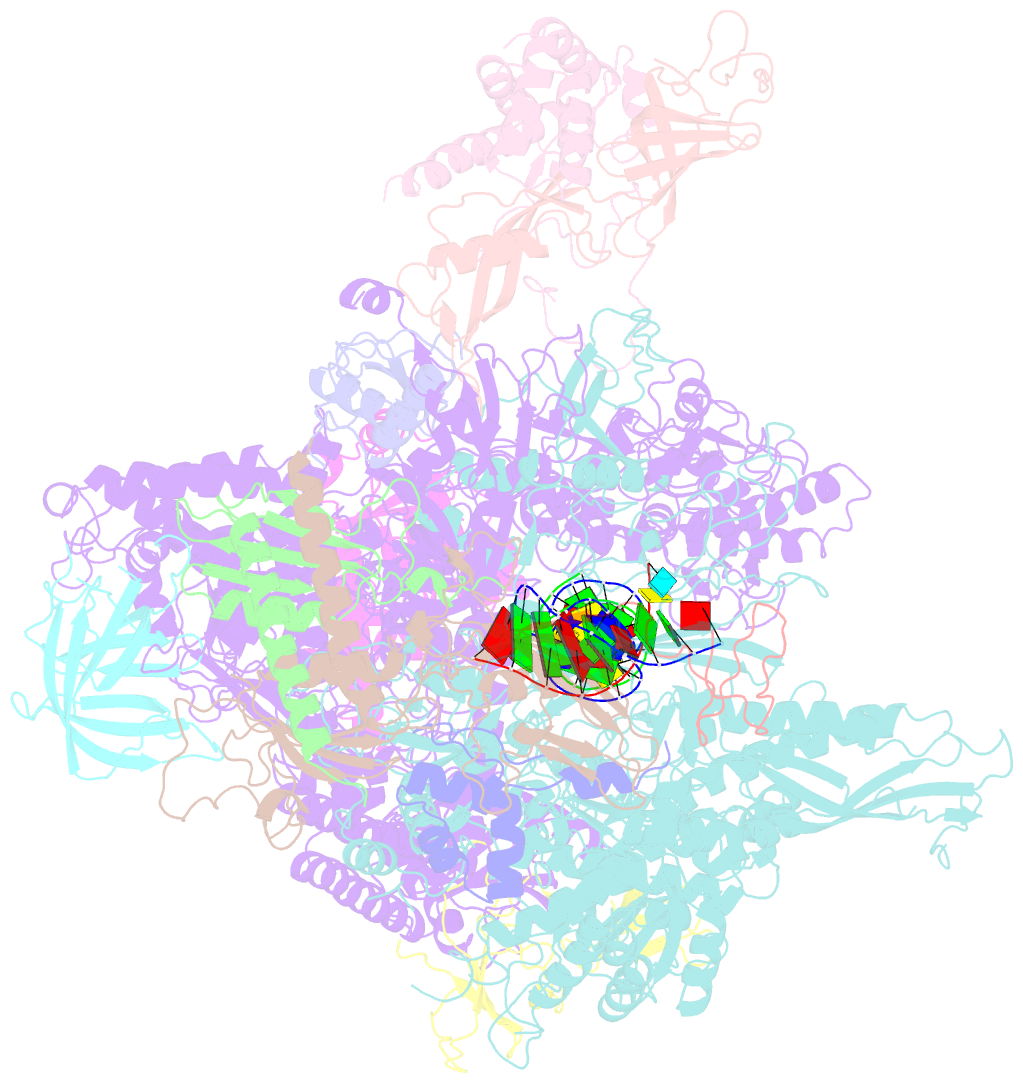
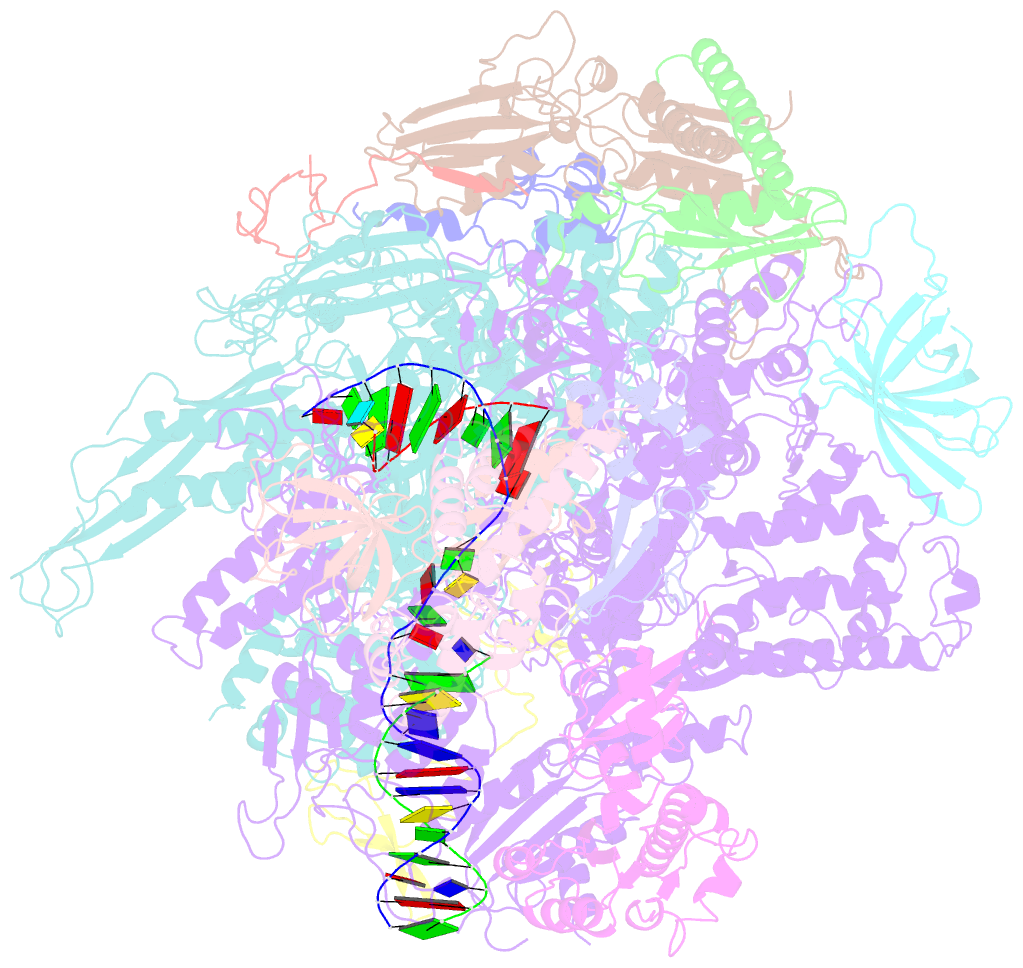
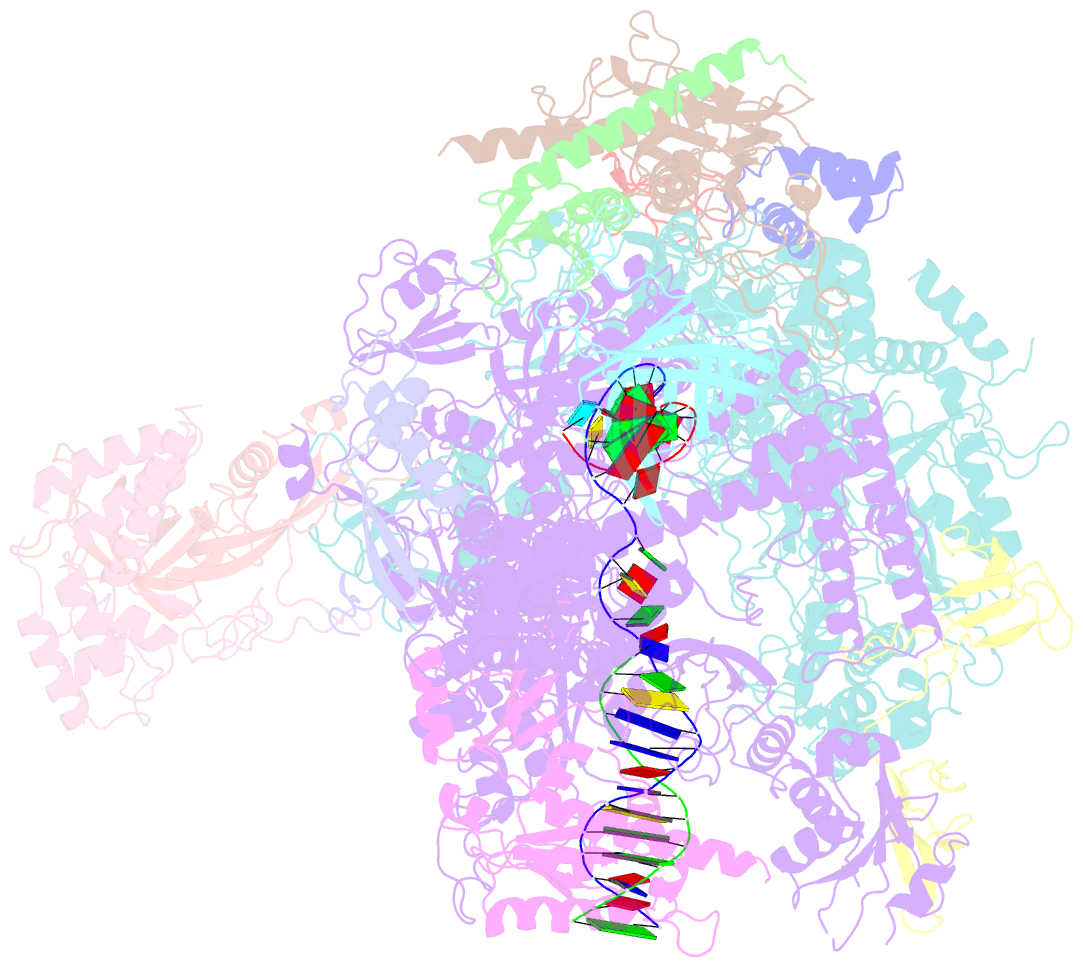
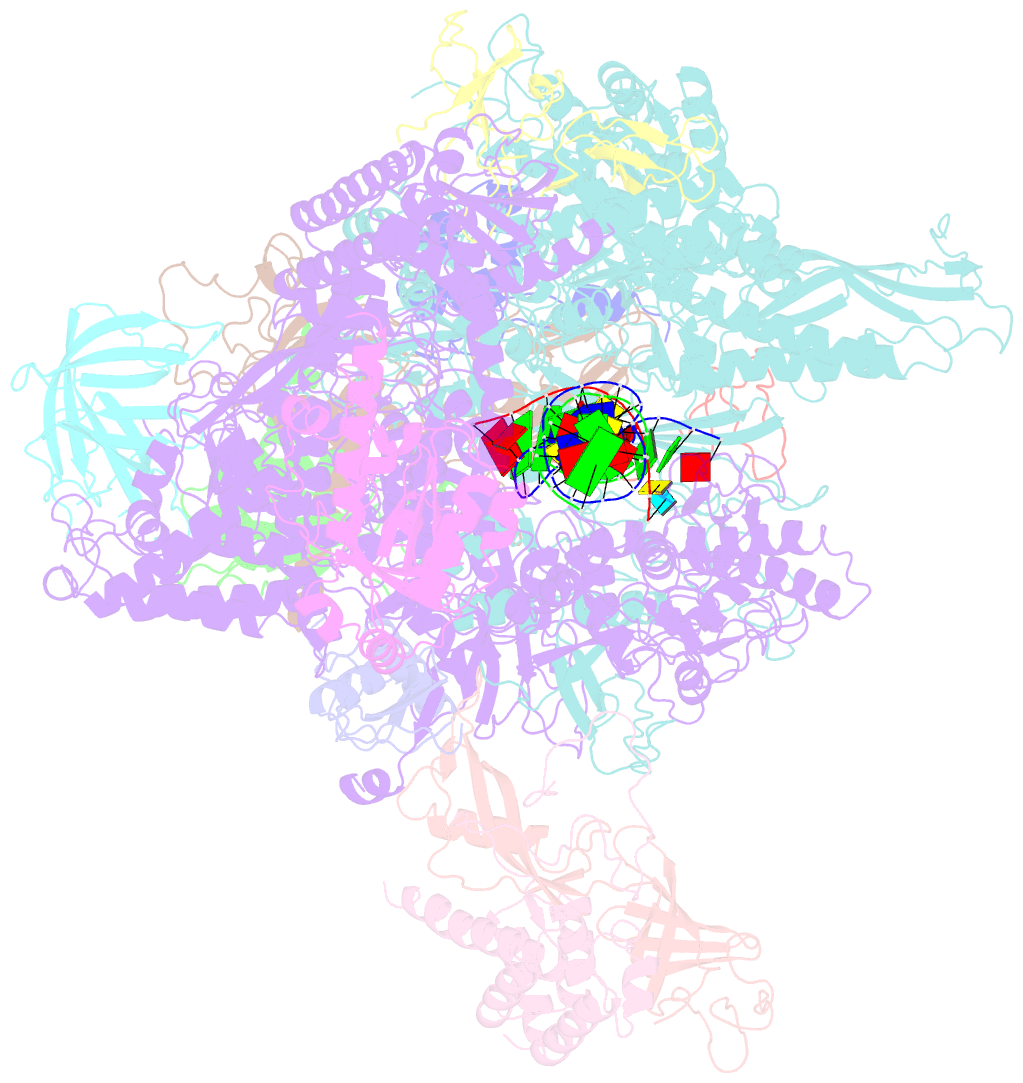
- Crystal structure of a transcribing RNA polymerase ii complex reveals a complete transcription bubble
- Reference
- Barnes CO, Calero M, Malik I, Graham BW, Spahr H, Lin G, Cohen AE, Brown IS, Zhang Q, Pullara F, Trakselis MA, Kaplan CD, Calero G (2015): "Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble." Mol.Cell, 59, 258-269. doi: 10.1016/j.molcel.2015.06.034.
- Abstract
- Notwithstanding numerous published structures of RNA Polymerase II (Pol II), structural details of Pol II engaging a complete nucleic acid scaffold have been lacking. Here, we report the structures of TFIIF-stabilized transcribing Pol II complexes, revealing the upstream duplex and full transcription bubble. The upstream duplex lies over a wedge-shaped loop from Rpb2 that engages its minor groove, providing part of the structural framework for DNA tracking during elongation. At the upstream transcription bubble fork, rudder and fork loop 1 residues spatially coordinate strand annealing and the nascent RNA transcript. At the downstream fork, a network of Pol II interactions with the non-template strand forms a rigid domain with the trigger loop (TL), allowing visualization of its open state. Overall, our observations suggest that "open/closed" conformational transitions of the TL may be linked to interactions with the non-template strand, possibly in a synchronized ratcheting manner conducive to polymerase translocation.