Summary information and primary citation
- PDB-id
- 5c9h; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (3.0 Å)
- Summary
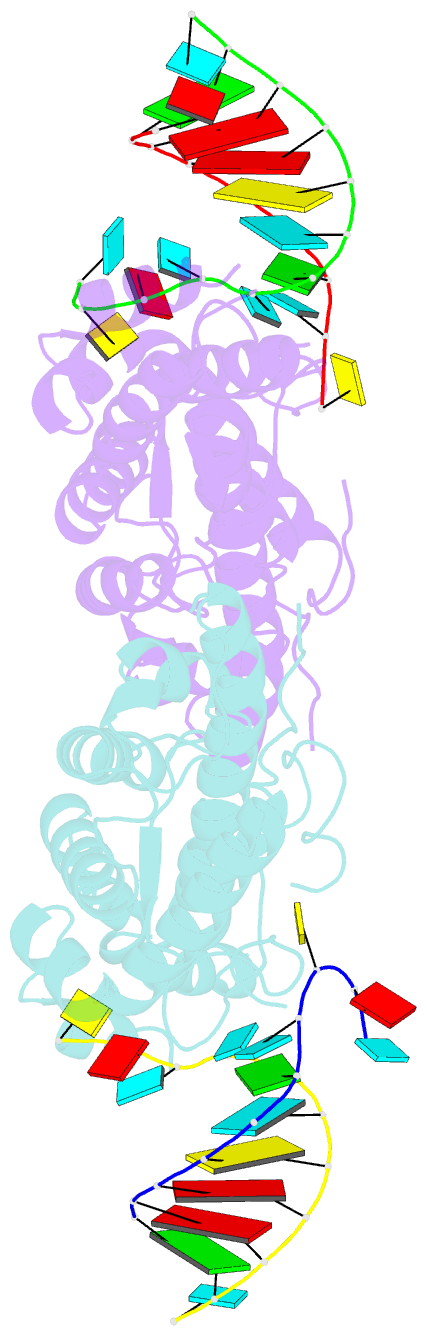
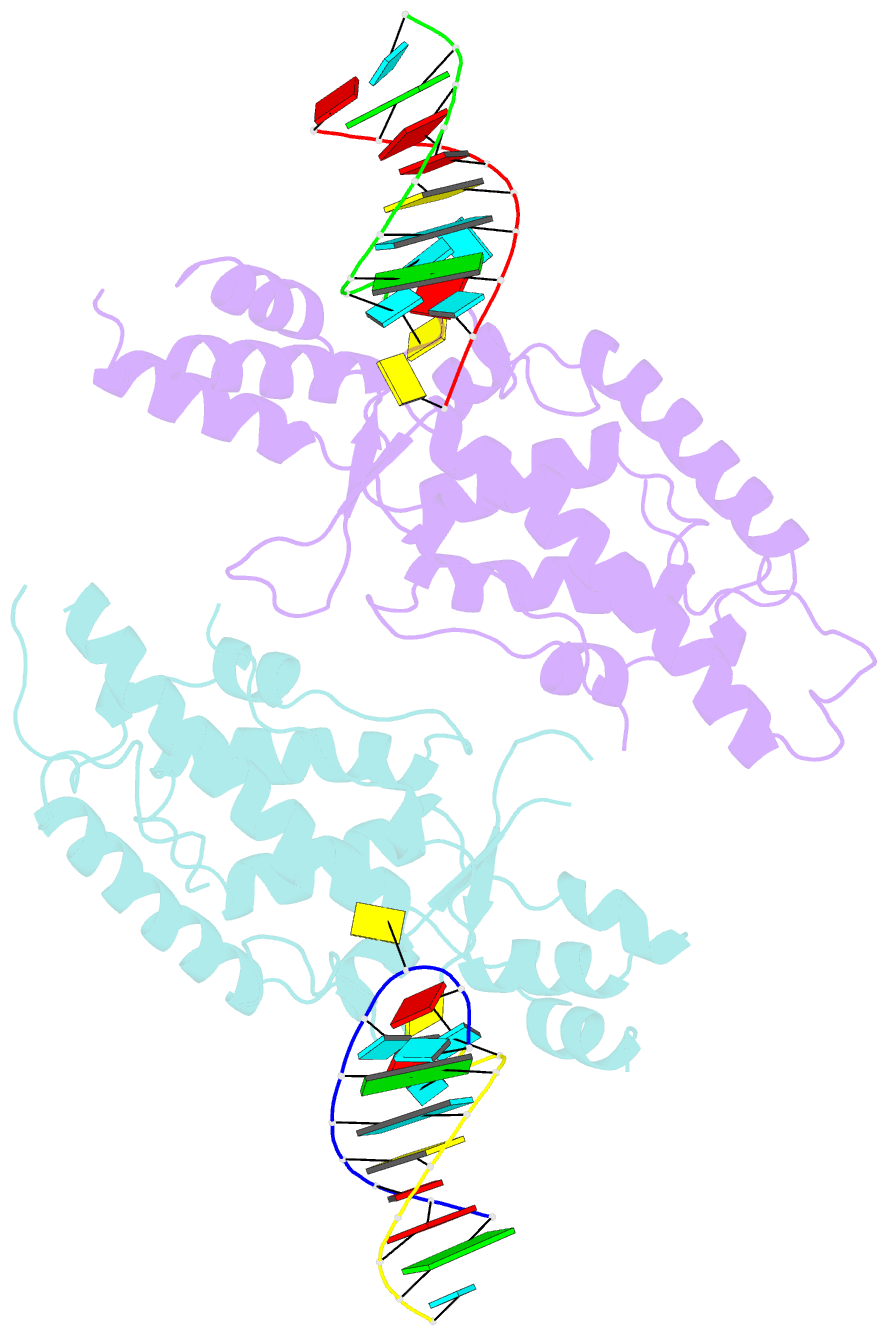
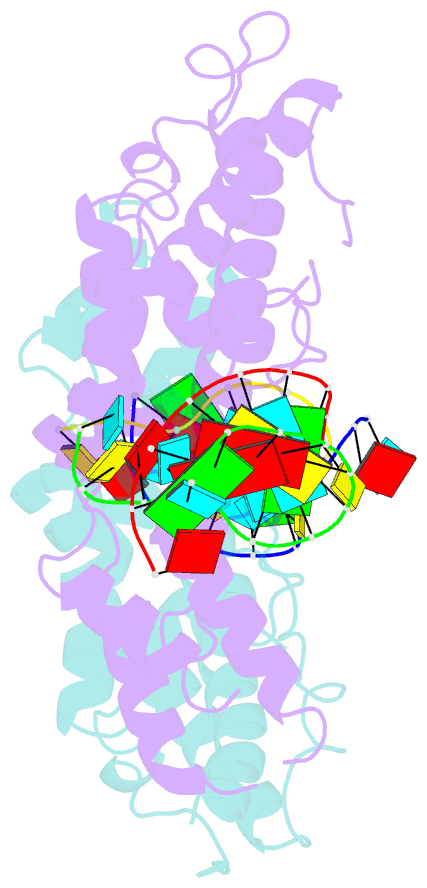
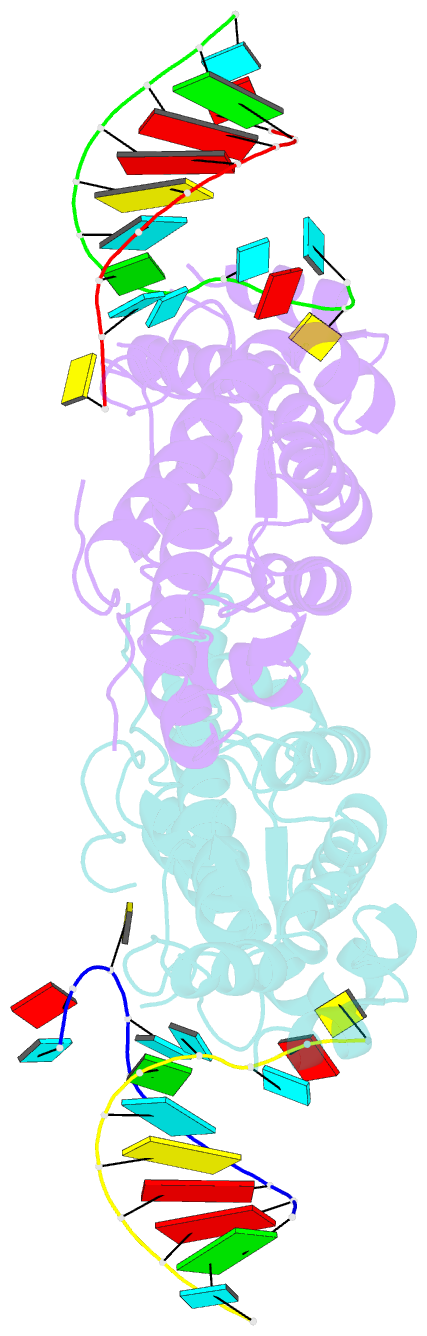
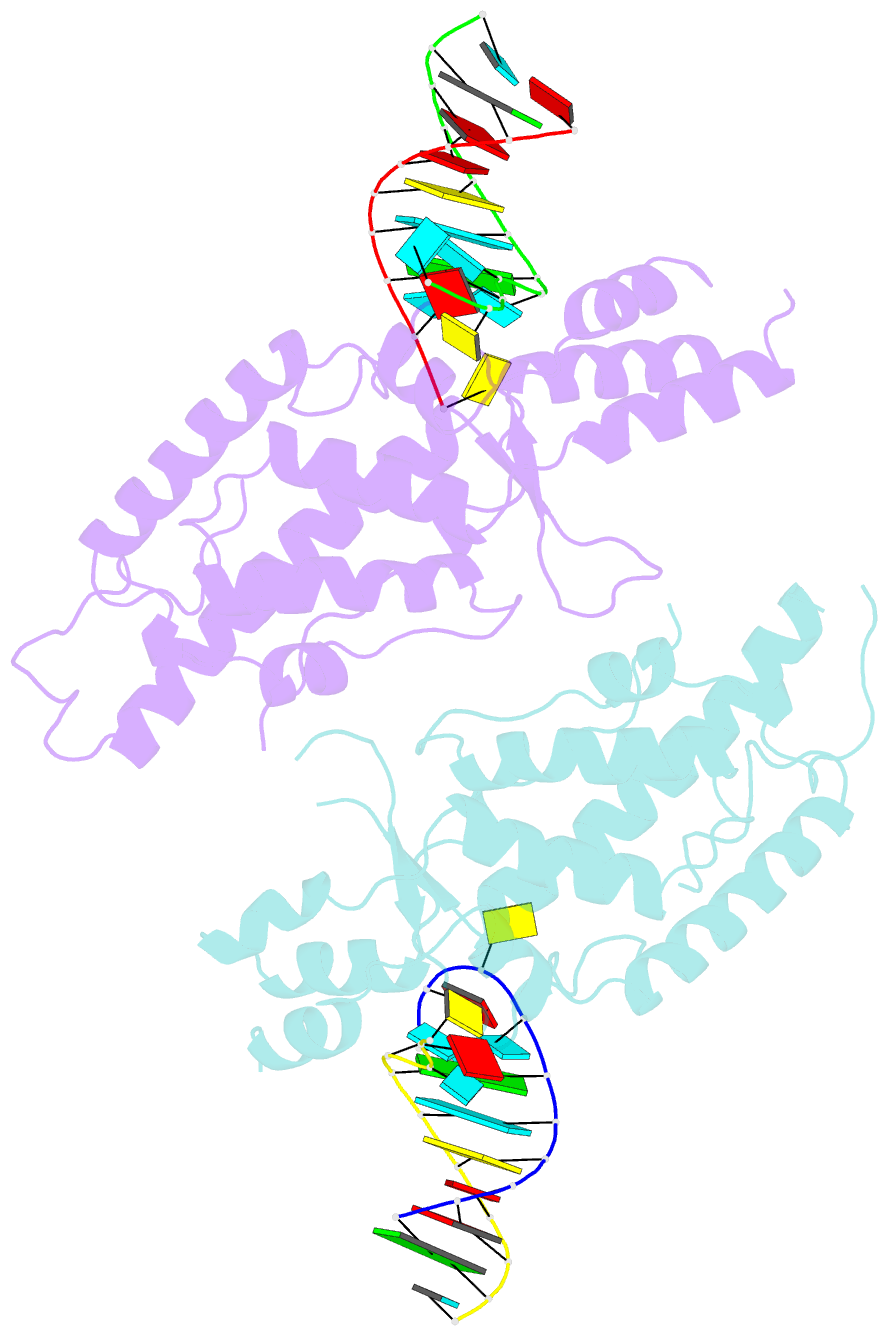
- Structural basis of template boundary definition in tetrahymena telomerase
- Reference
- Jansson LI, Akiyama BM, Ooms A, Lu C, Rubin SM, Stone MD (2015): "Structural basis of template-boundary definition in Tetrahymena telomerase." Nat.Struct.Mol.Biol., 22, 883-888. doi: 10.1038/nsmb.3101.
- Abstract
- Telomerase is required to maintain repetitive G-rich telomeric DNA sequences at chromosome ends. To do so, the telomerase reverse transcriptase (TERT) subunit reiteratively uses a small region of the integral telomerase RNA (TER) as a template. An essential feature of telomerase catalysis is the strict definition of the template boundary to determine the precise TER nucleotides to be reverse transcribed by TERT. We report the 3-Å crystal structure of the Tetrahymena TERT RNA-binding domain (tTRBD) bound to the template boundary element (TBE) of TER. tTRBD is wedged into the base of the TBE RNA stem-loop, and each of the flanking RNA strands wraps around opposite sides of the protein domain. The structure illustrates how the tTRBD establishes the template boundary by positioning the TBE at the correct distance from the TERT active site to prohibit copying of nontemplate nucleotides.