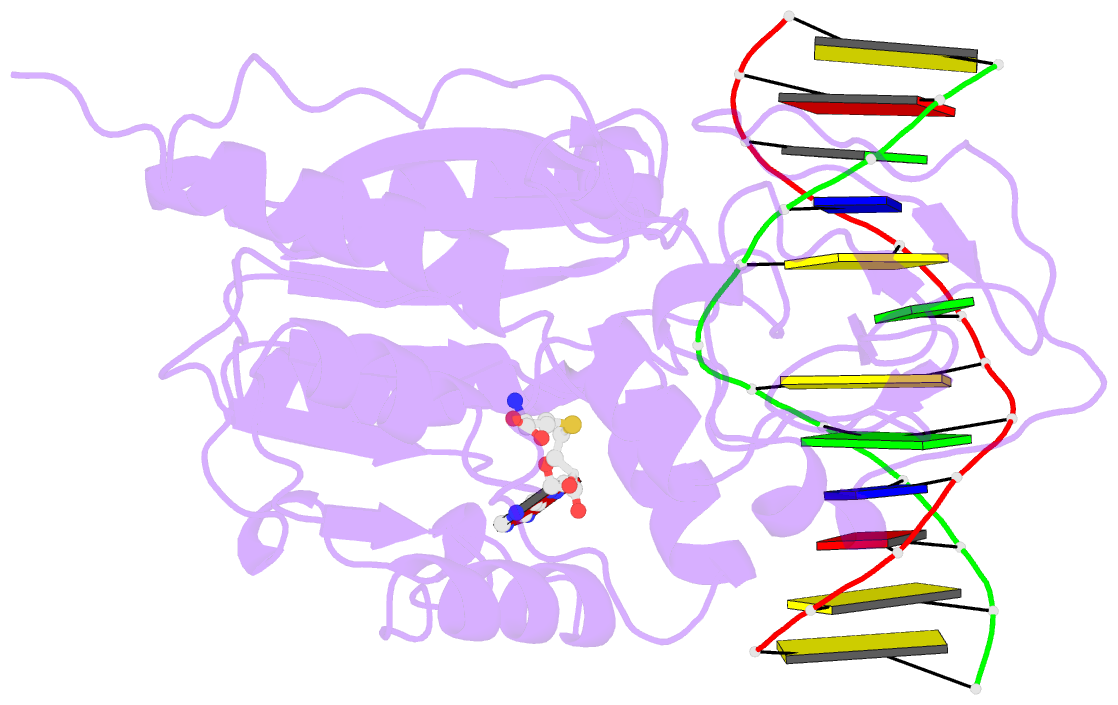
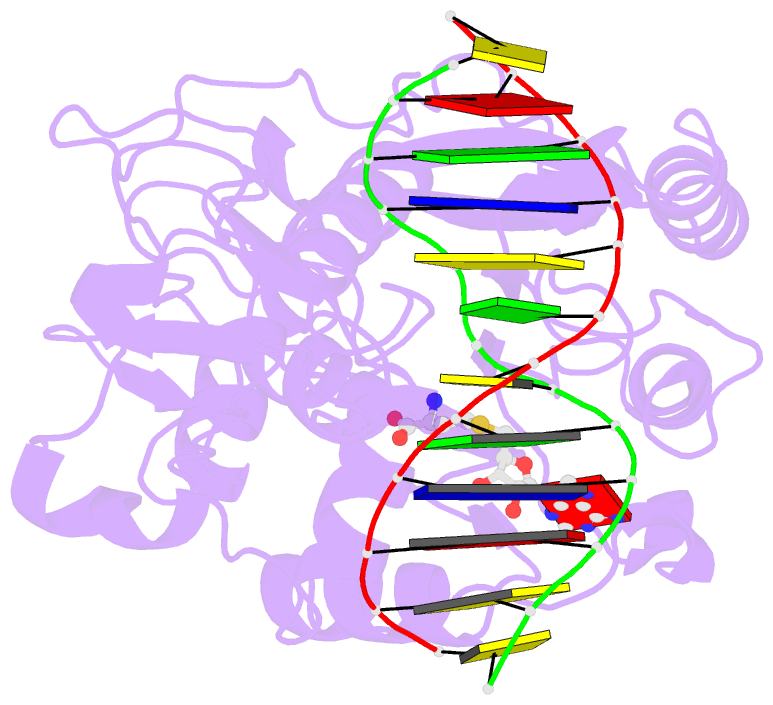
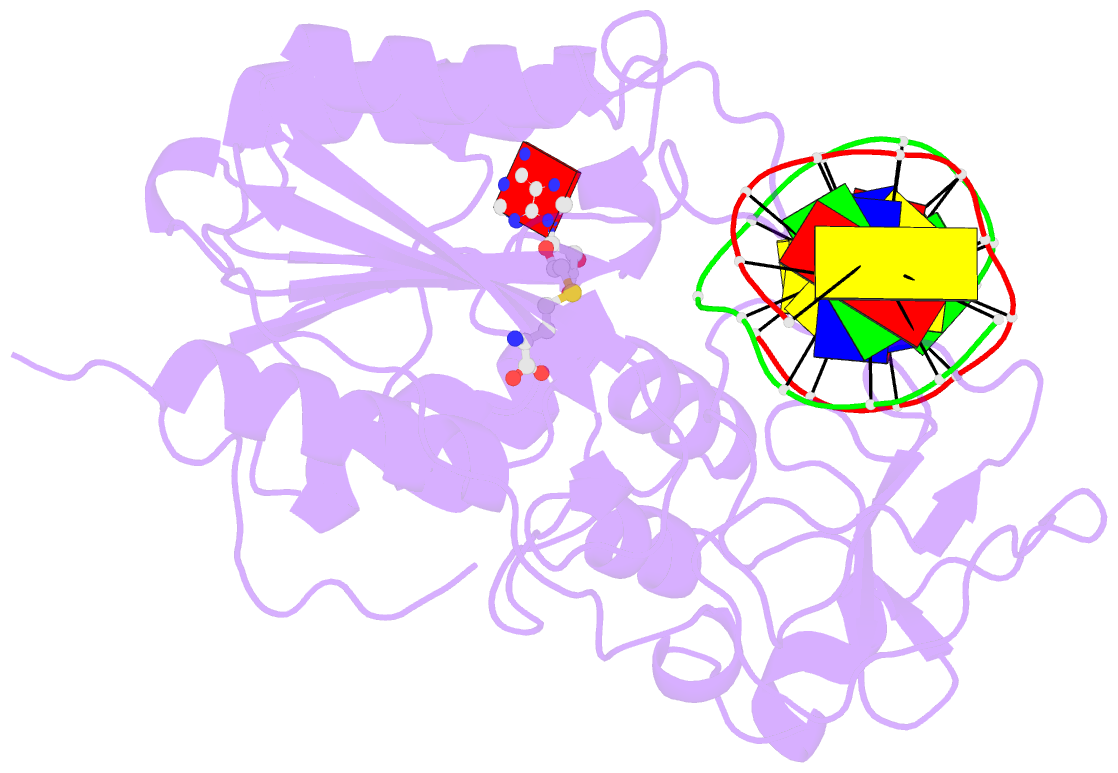
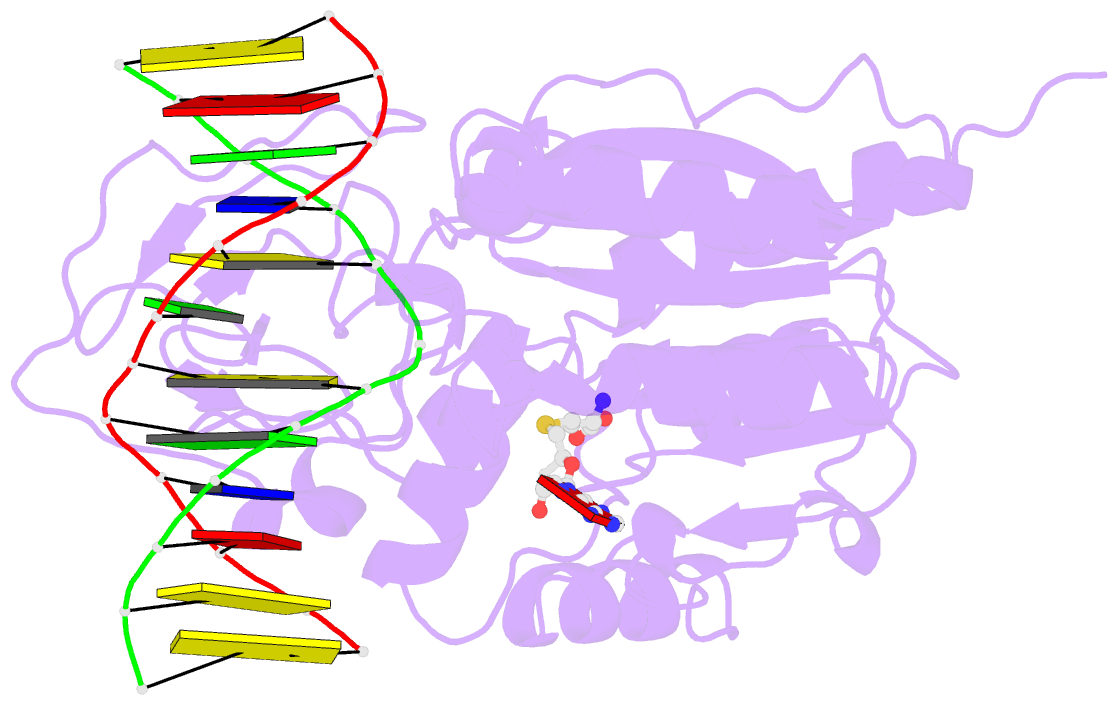
Summary information and primary citation
- PDB-id
- 5ciy; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transferase-DNA
- Method
- X-ray (1.594 Å)
- Summary
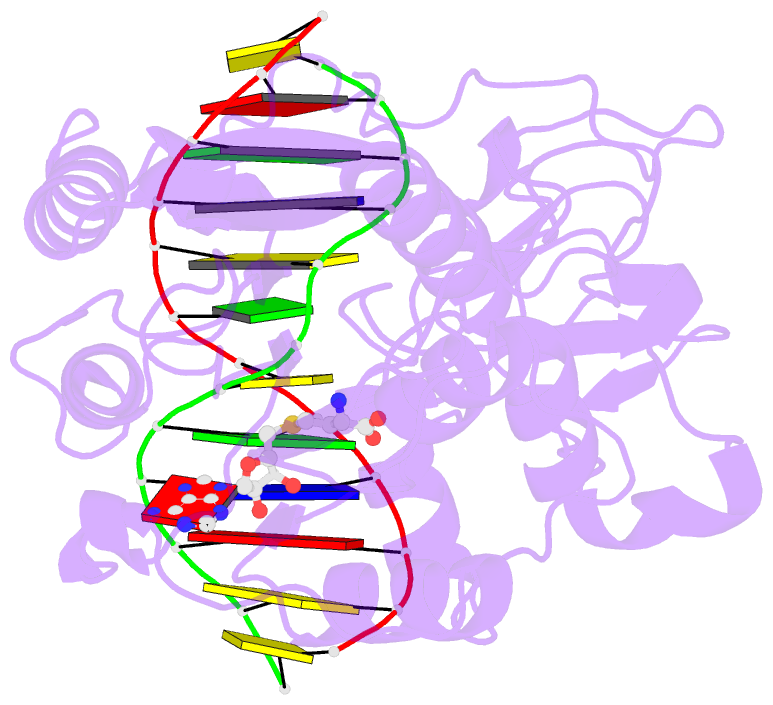
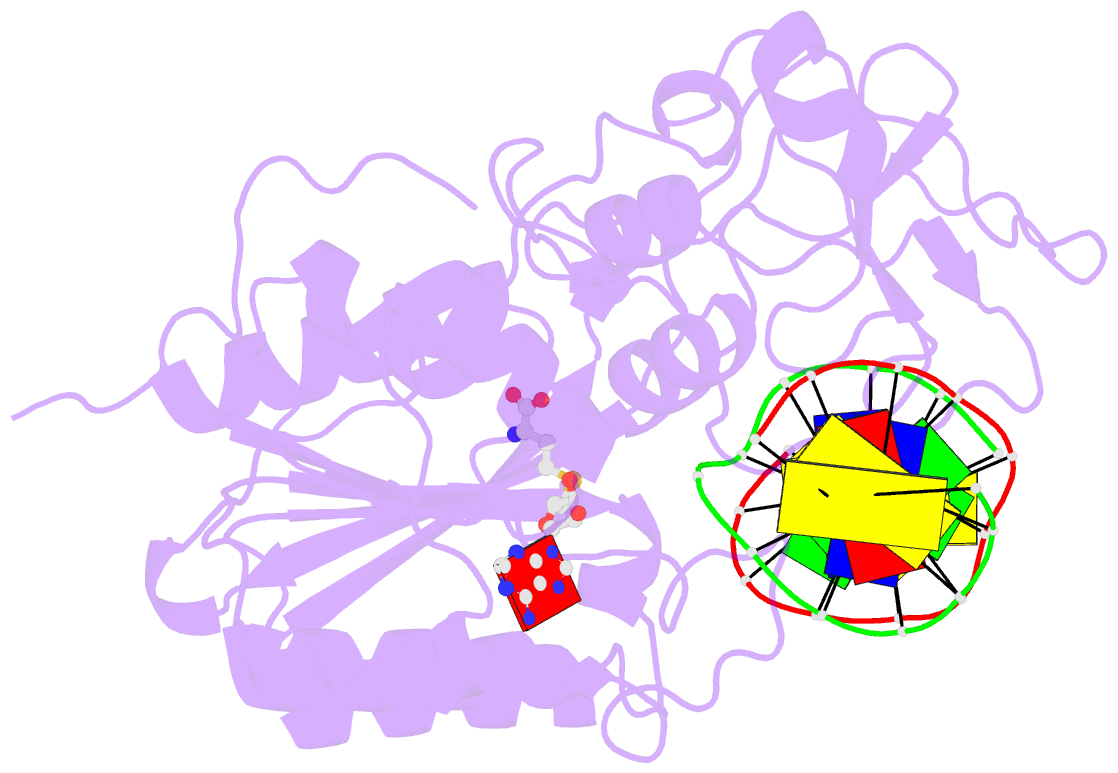
- Structural basis of the recognition of h3k36me3 by dnmt3b pwwp domain
- Reference
- Rondelet G, Dal Maso T, Willems L, Wouters J (2016): "Structural basis for recognition of histone H3K36me3 nucleosome by human de novo DNA methyltransferases 3A and 3B." J.Struct.Biol., 194, 357-367. doi: 10.1016/j.jsb.2016.03.013.
- Abstract
- DNA methylation is an important epigenetic modification involved in chromatin organization and gene expression. The function of DNA methylation depends on cell context and is correlated with histone modification patterns. In particular, trimethylation of Lys36 on histone H3 tail (H3K36me3) is associated with DNA methylation and elongation phase of transcription. PWWP domains of the de novo DNA methyltransferases DNMT3A and DNMT3B read this epigenetic mark to guide DNA methylation. Here we report the first crystal structure of the DNMT3B PWWP domain-H3K36me3 complex. Based on this structure, we propose a model of the DNMT3A PWWP domain-H3K36me3 complex and build a model of DNMT3A (PWWP-ADD-CD) in a nucleosomal context. The trimethylated side chain of Lys36 (H3K36me3) is inserted into an aromatic cage similar to the "Royal" superfamily domains known to bind methylated histones. A key interaction between trimethylated Lys36 and a conserved water molecule stabilized by Ser270 explains the lack of affinity of mutated DNMT3B (S270P) for the H3K36me3 epigenetic mark in the ICF (Immunodeficiency, Centromeric instability and Facial abnormalities) syndrome. The model of the DNMT3A-DNMT3L heterotetramer in complex with a dinucleosome highlights the mechanism for recognition of nucleosome by DNMT3s and explains the periodicity of de novo DNA methylation.