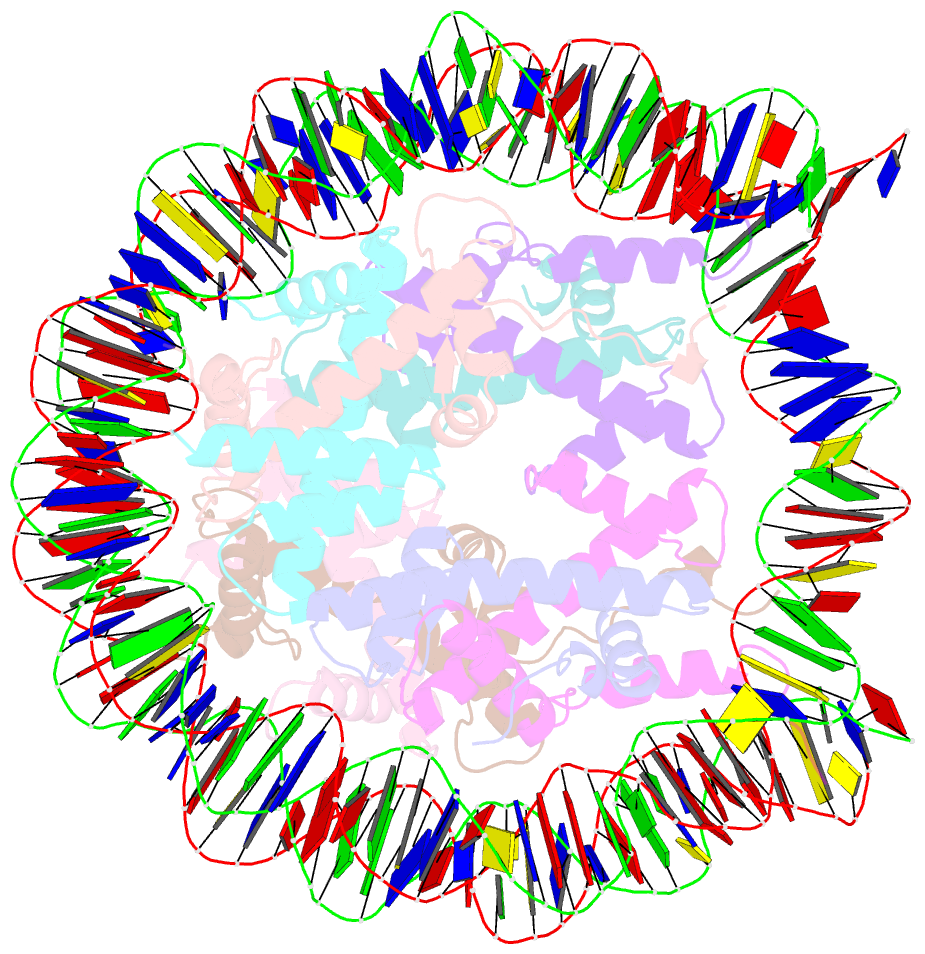
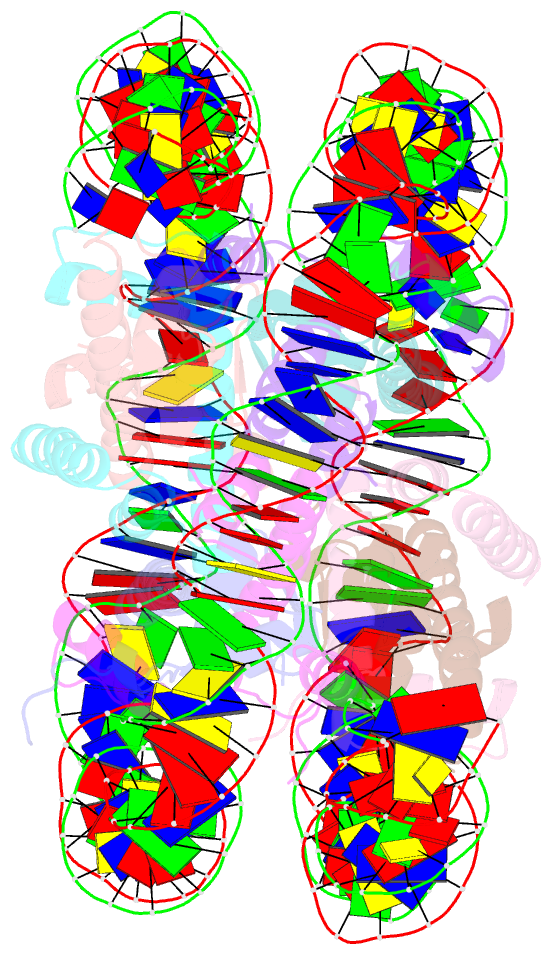
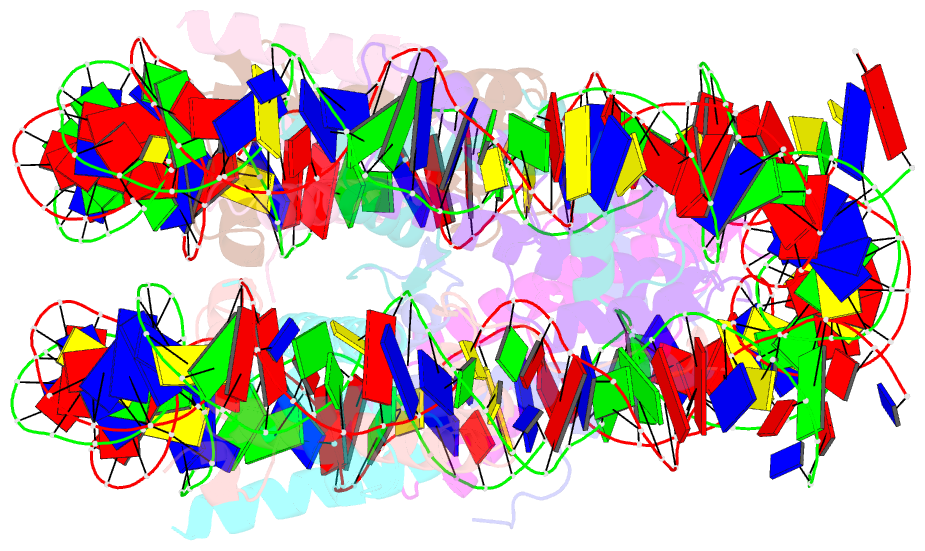
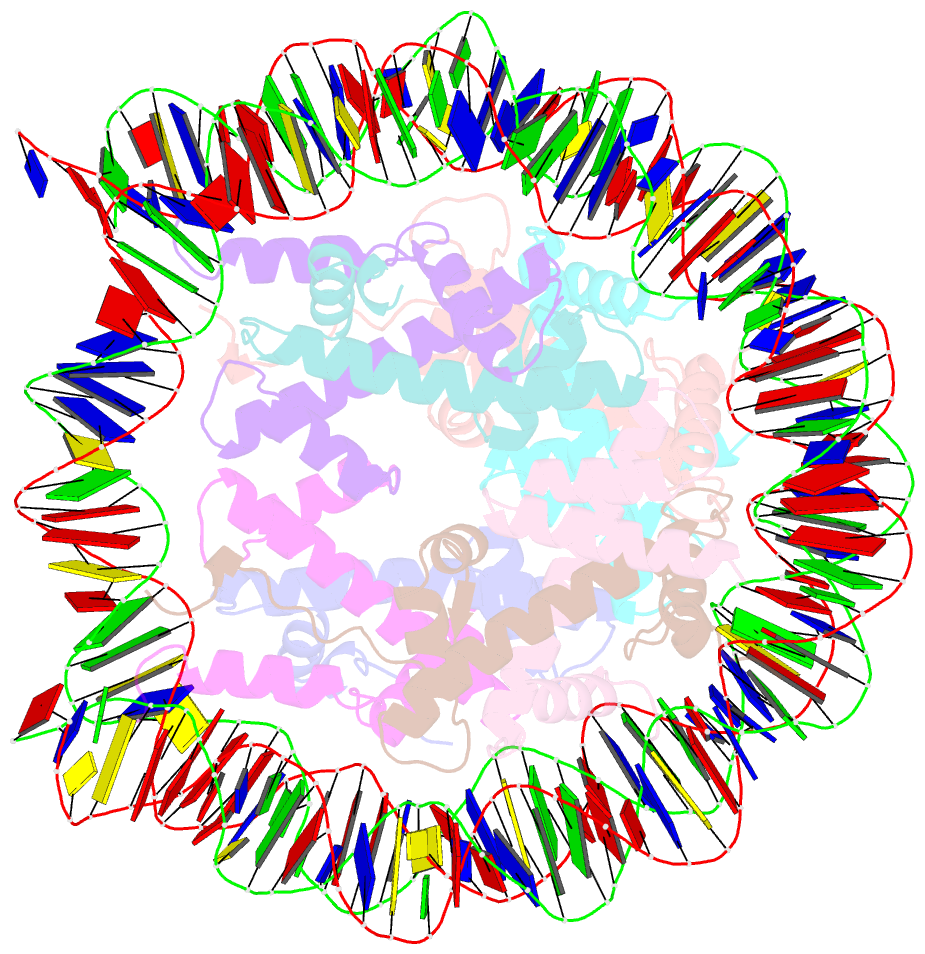
Summary information and primary citation
- PDB-id
- 5cpj; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- "structural protein-DNA
- Method
- X-ray (3.15 Å)
- Summary
- Nucleosome containing methylated sat2r DNA
- Reference
- Osakabe A, Adachi F, Arimura Y, Maehara K, Ohkawa Y, Kurumizaka H (2015): "Influence of DNA methylation on positioning and DNA flexibility of nucleosomes with pericentric satellite DNA." Open Biology, 5. doi: 10.1098/rsob.150128.
- Abstract
- DNA methylation occurs on CpG sites and is important to form pericentric heterochromatin domains. The satellite 2 sequence, containing seven CpG sites, is located in the pericentric region of human chromosome 1 and is highly methylated in normal cells. In contrast, the satellite 2 region is reportedly hypomethylated in cancer cells, suggesting that the methylation status may affect the chromatin structure around the pericentric regions in tumours. In this study, we mapped the nucleosome positioning on the satellite 2 sequence in vitro and found that DNA methylation modestly affects the distribution of the nucleosome positioning. The micrococcal nuclease assay revealed that the DNA end flexibility of the nucleosomes changes, depending on the DNA methylation status. However, the structures and thermal stabilities of the nucleosomes are unaffected by DNA methylation. These findings provide new information to understand how DNA methylation functions in regulating pericentric heterochromatin formation and maintenance in normal and malignant cells.