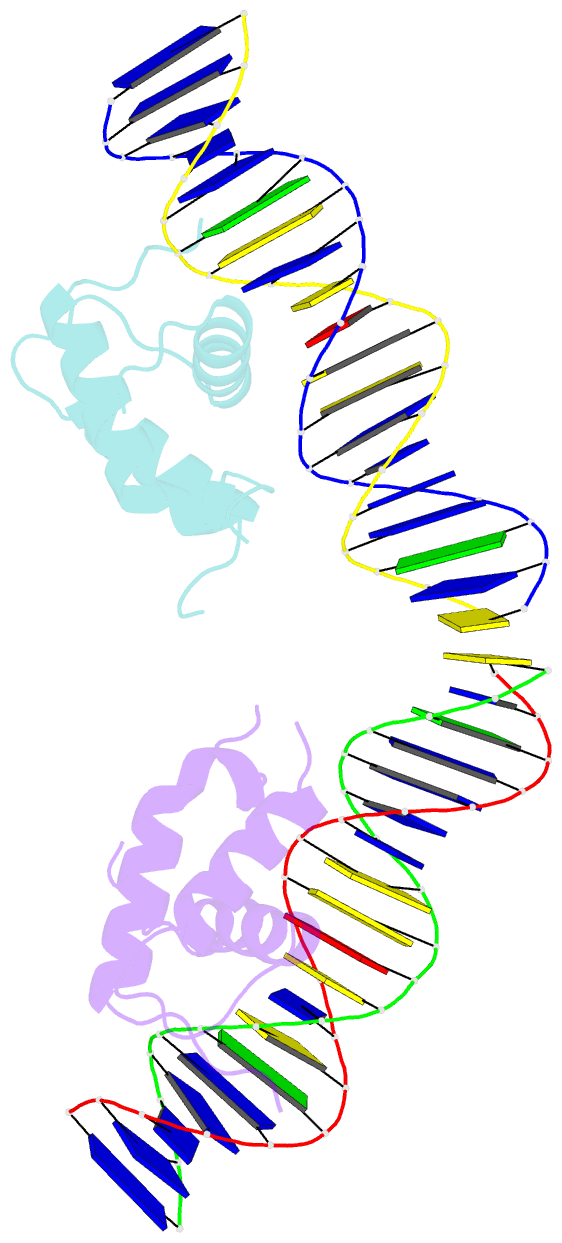
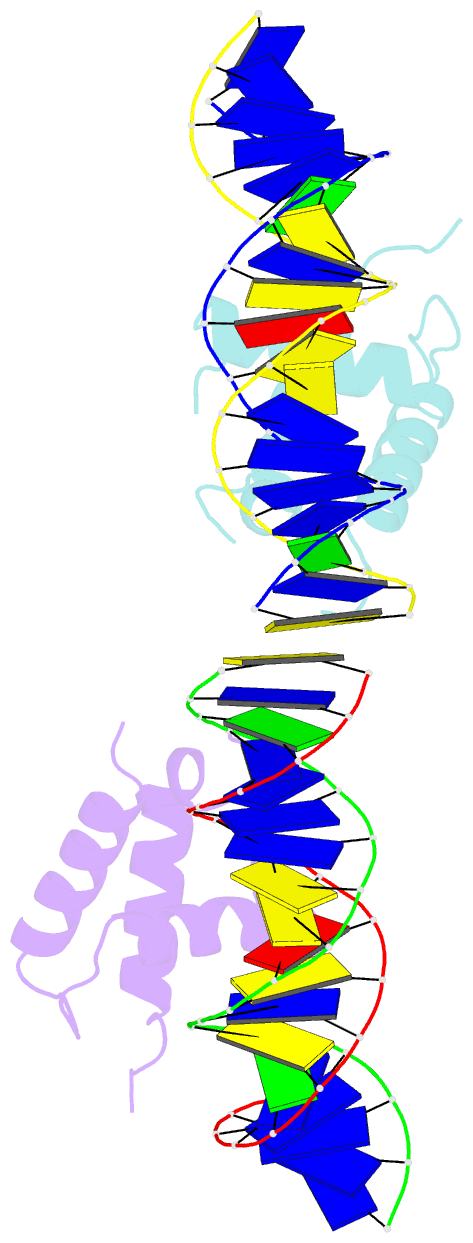
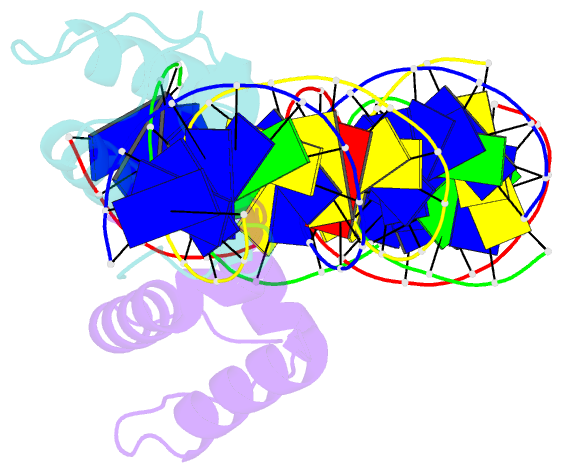
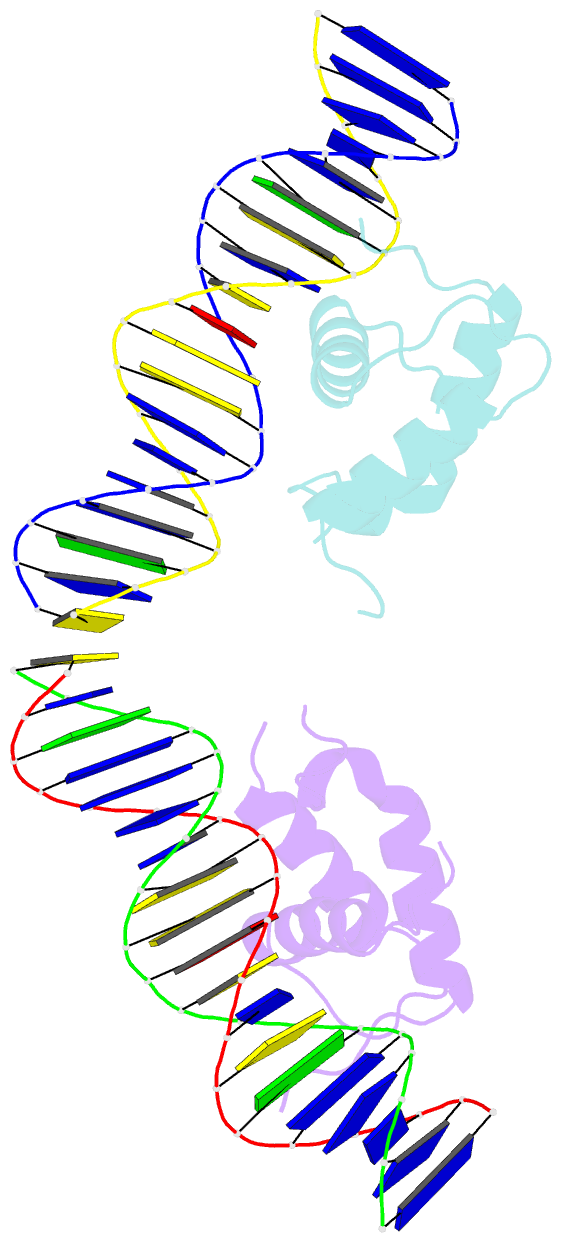
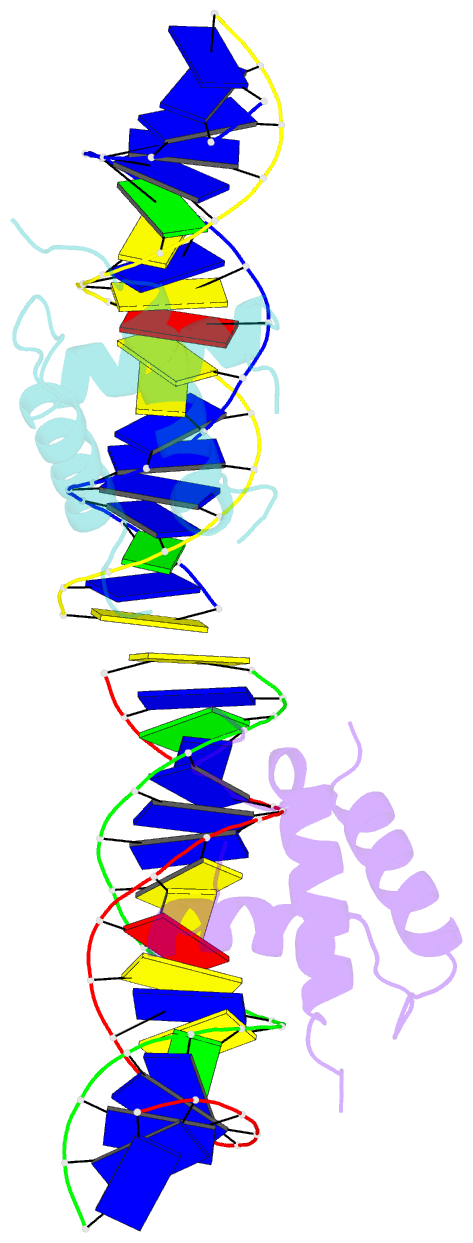
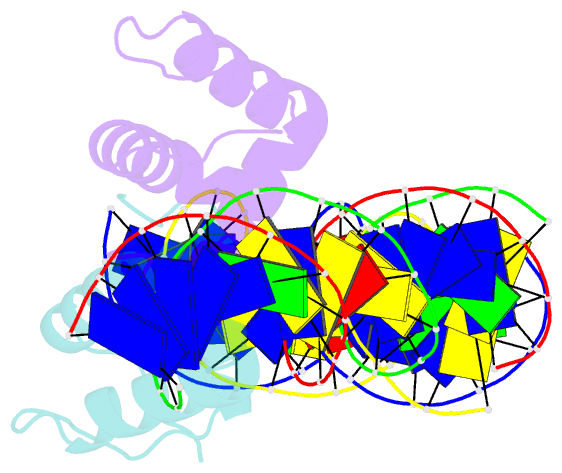
Summary information and primary citation
- PDB-id
- 5cqq; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (3.1 Å)
- Summary
- Crystal structure of the drosophila zeste DNA binding domain in complex with DNA
- Reference
- Gao GN, Wang M, Yang N, Huang Y, Xu RM (2015): "Structure of Zeste-DNA Complex Reveals a New Modality of DNA Recognition by Homeodomain-Like Proteins." J.Mol.Biol., 427, 3824-3833. doi: 10.1016/j.jmb.2015.10.008.
- Abstract
- Drosophila Zeste is a DNA binding protein important for chromatin-targeted regulation of gene expression. It is best studied in the context of transvection-a mechanism of interallelic gene regulation involving paired chromosomes-and repression of the expression of white by Zeste mutants. Both of these functions depend on the DNA binding and self-association properties of Zeste, but the underlying structural basis remains unknown. Here we report the crystal structure of the DNA binding domain of Zeste in complex with a 19-bp DNA duplex containing the consensus recognition sequence motif. The structure reveals a helix-turn-helix Myb/homeodomain-like fold with the Zeste-specific insertion sequence forming a short helix and a long loop. Direct base contacts by the major groove binding helix principally account for the sequence-specific recognition, and backbone contacts via the Zeste-specific insertion are mainly responsible for the length requirement and the orientation of DNA. Our structural and biochemical characterizations of the DNA binding property of Zeste uncover an altered DNA binding modality of homeodomain-like proteins, and the structural information should facilitate the unraveling of the intricate mechanism of Zeste in regulation of gene expression.