Summary information and primary citation
- PDB-id
- 5crx; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- protein-DNA
- Method
- X-ray (2.7 Å)
- Summary
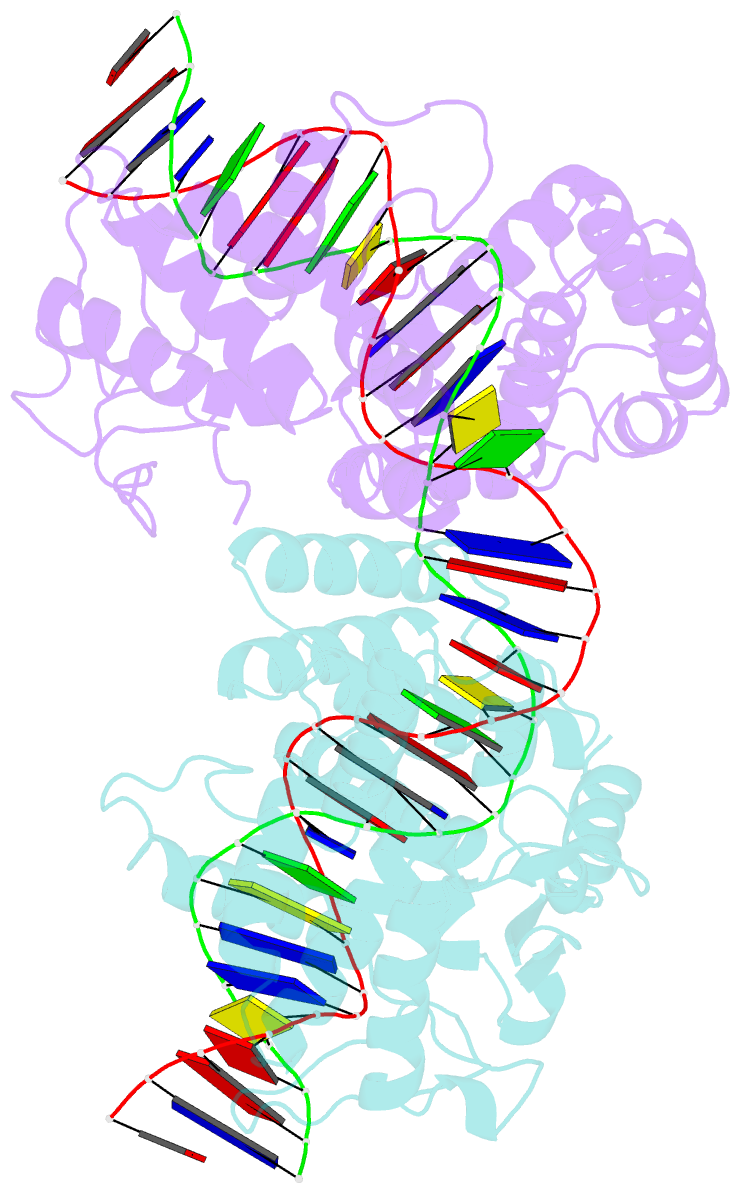
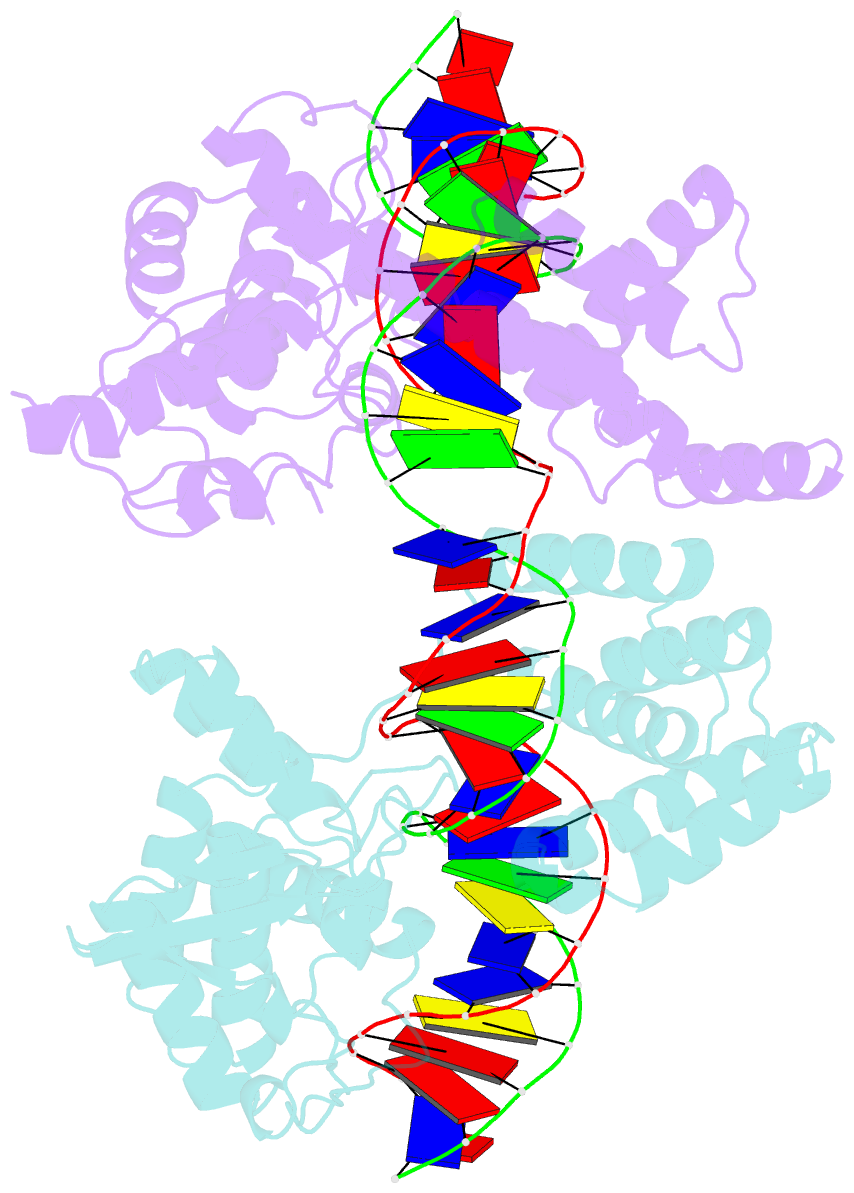
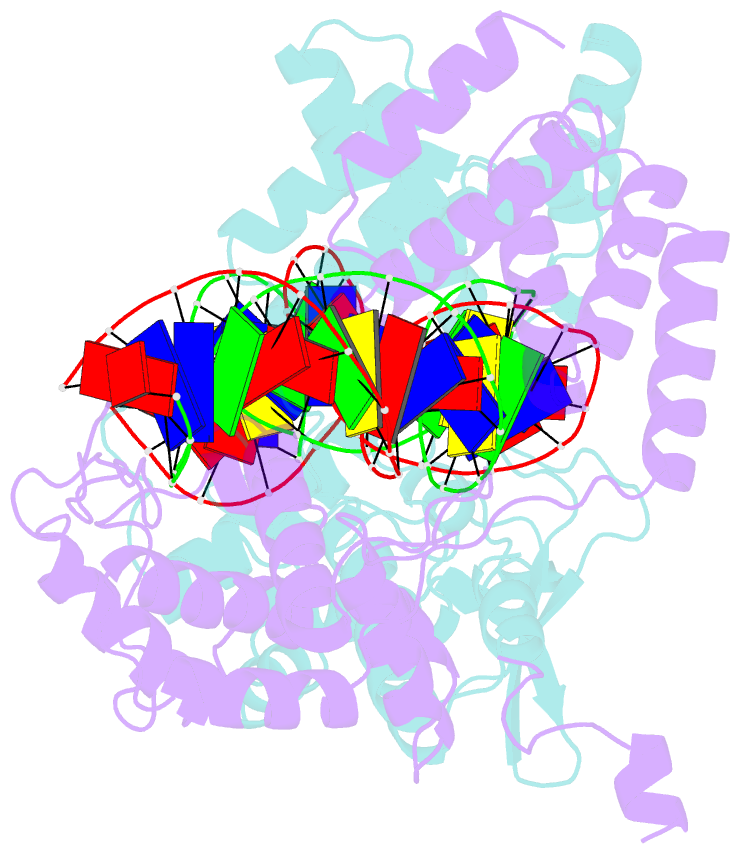
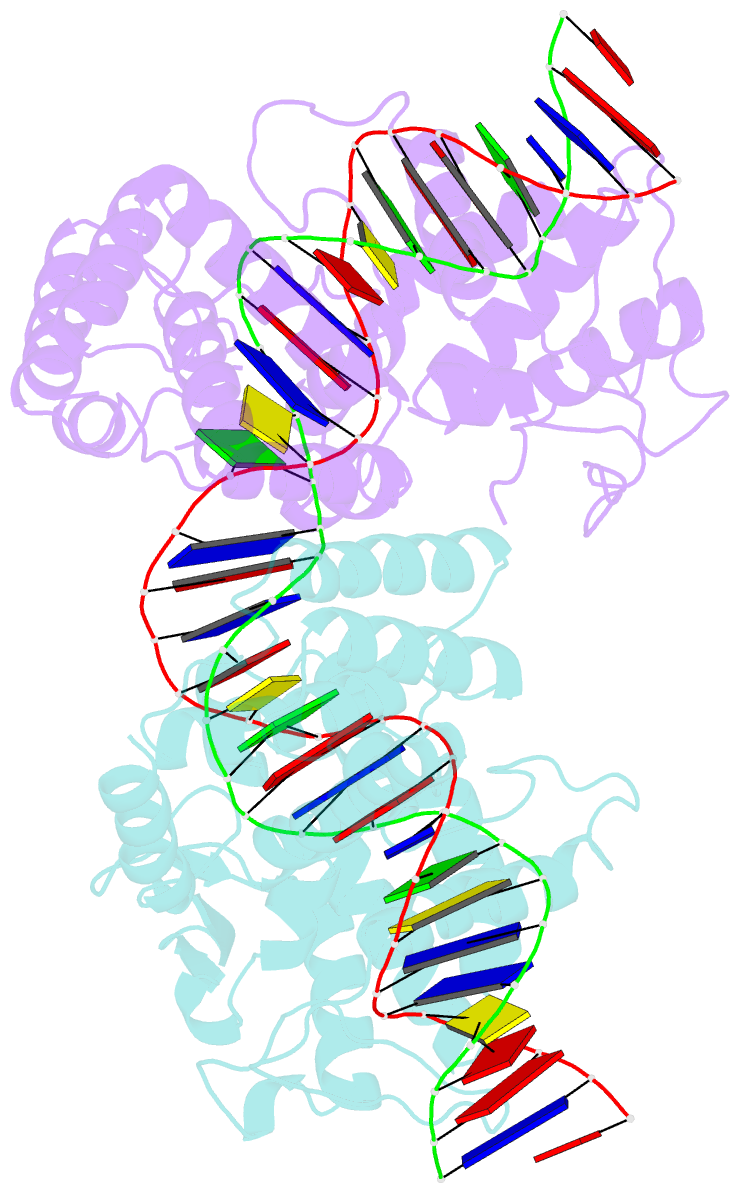
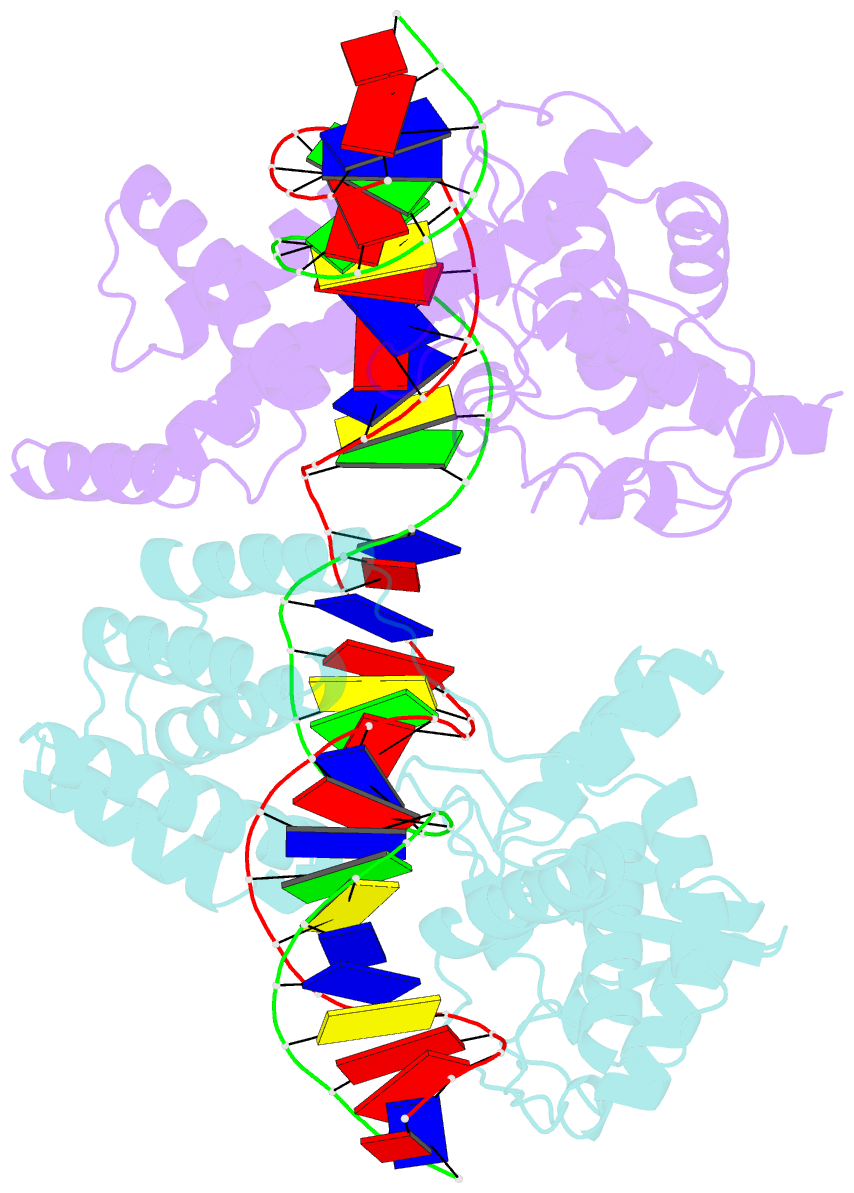
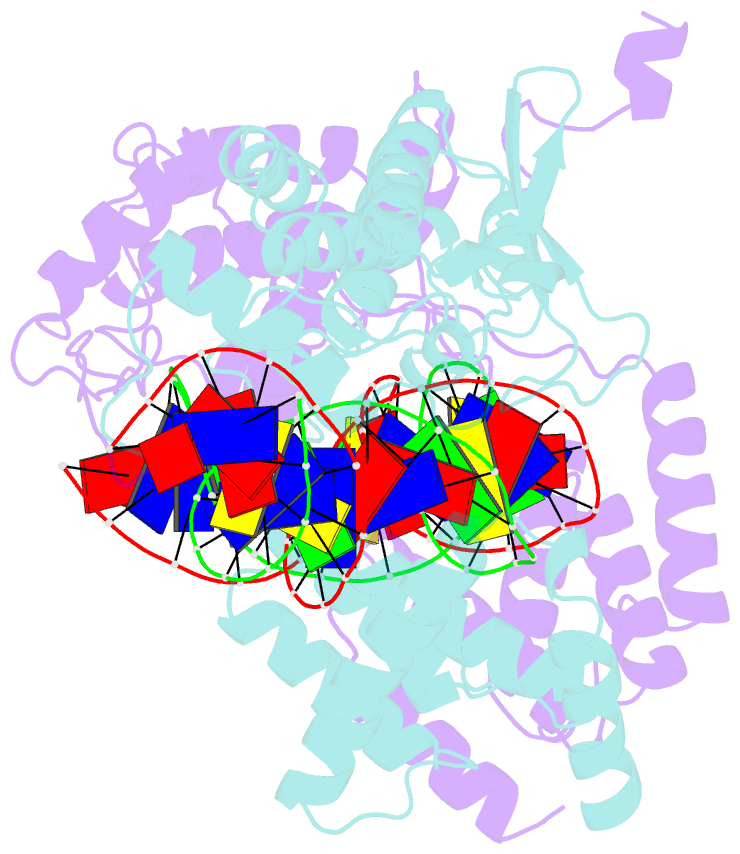
- Asymmetric DNA-bending in the cre-loxp site-specific recombination synapse
- Reference
- Guo F, Gopaul DN, Van Duyne GD (1999): "Asymmetric DNA bending in the Cre-loxP site-specific recombination synapse." Proc.Natl.Acad.Sci.USA, 96, 7143-7148. doi: 10.1073/pnas.96.13.7143.
- Abstract
- Cre recombinase catalyzes site-specific recombination between two 34-bp loxP sites in a variety of DNA substrates. At the start of the recombination pathway, the loxP sites are each bound by two recombinase molecules, and synapsis of the sites is mediated by Cre-Cre interactions. We describe the structures of synaptic complexes formed between a symmetrized loxP site and two Cre mutants that are defective in strand cleavage. The DNA in these complexes is bent sharply at a single base pair step at one end of the crossover region in a manner that is atypical of protein-induced DNA bends. A large negative roll (-49 degrees) and a positive tilt (16 degrees) open the major groove toward the center of the synapse and compress the minor groove toward the protein-DNA interface. The bend direction of the site appears to determine which of the two DNA substrate strands will be cleaved and exchanged in the initial stages of the recombination pathway. These results provide a structural basis for the observation that exchange of DNA strands proceeds in a defined order in some tyrosine recombinase systems. The Cre-loxS synaptic complex structure supports a model in which synapsis of the loxP sites results in formation of a Holliday junction-like DNA architecture that is maintained through the initial cleavage and strand exchange steps in the site-specific recombination pathway.