Summary information and primary citation
- PDB-id
- 5d23; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA
- Method
- X-ray (1.95 Å)
- Summary
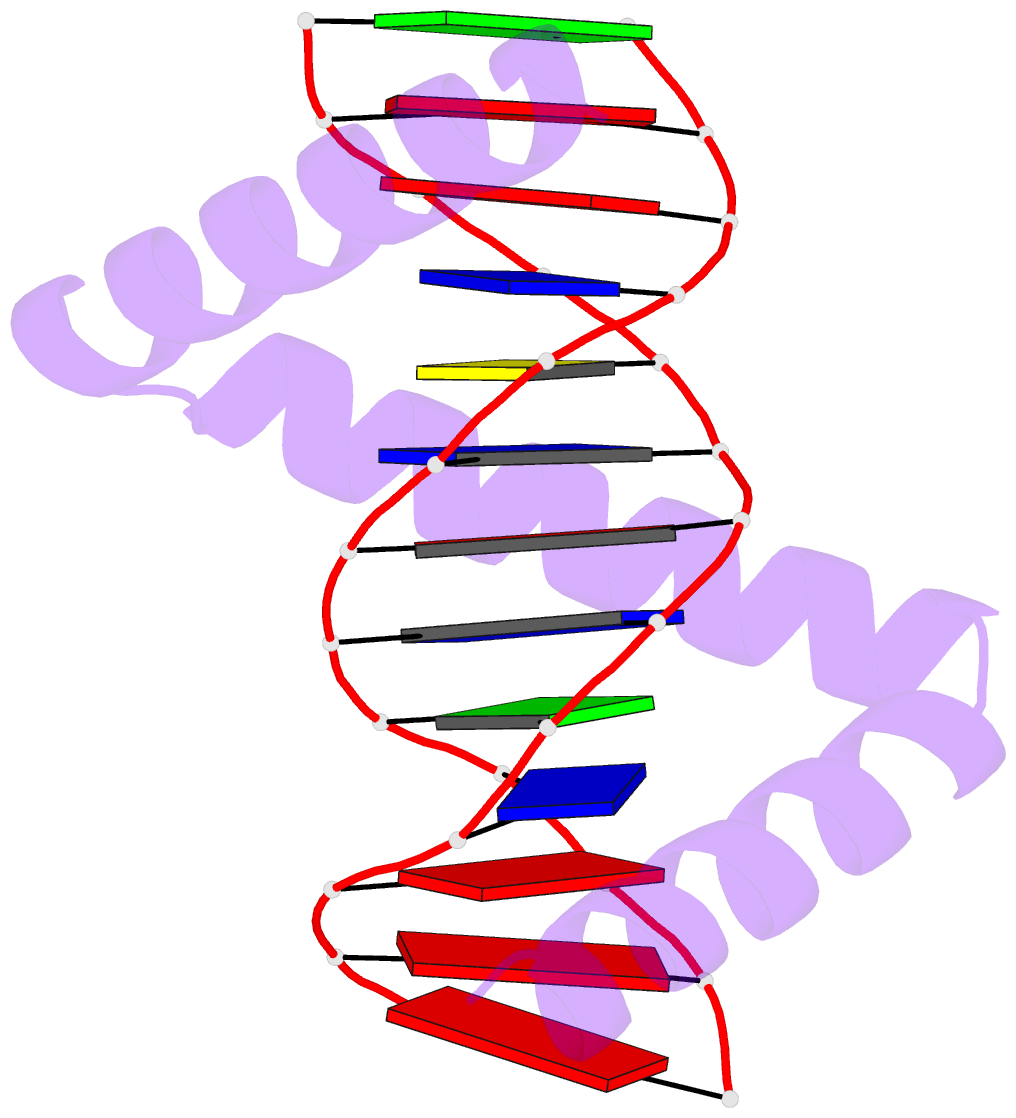
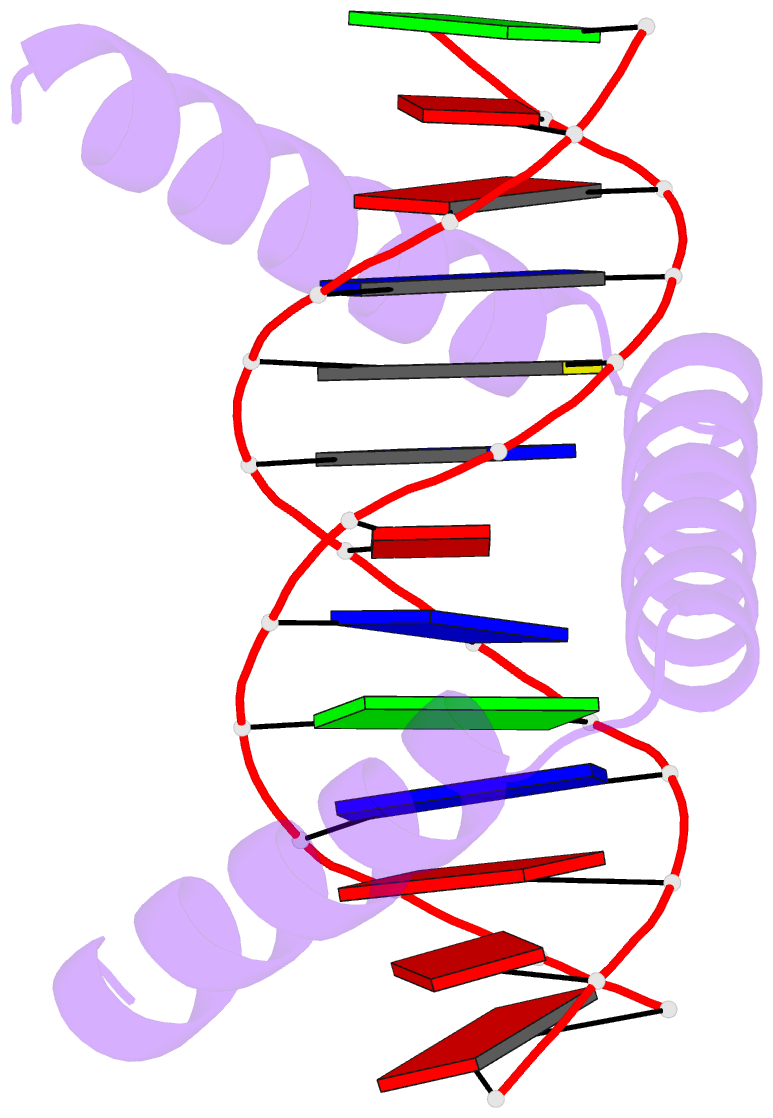
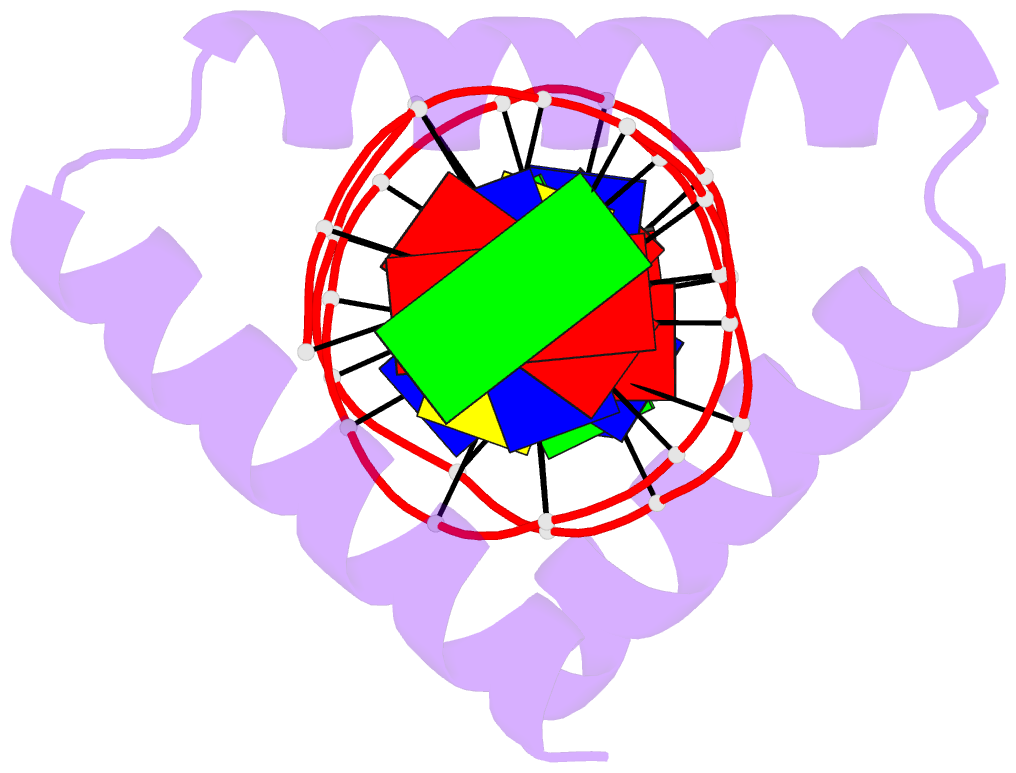
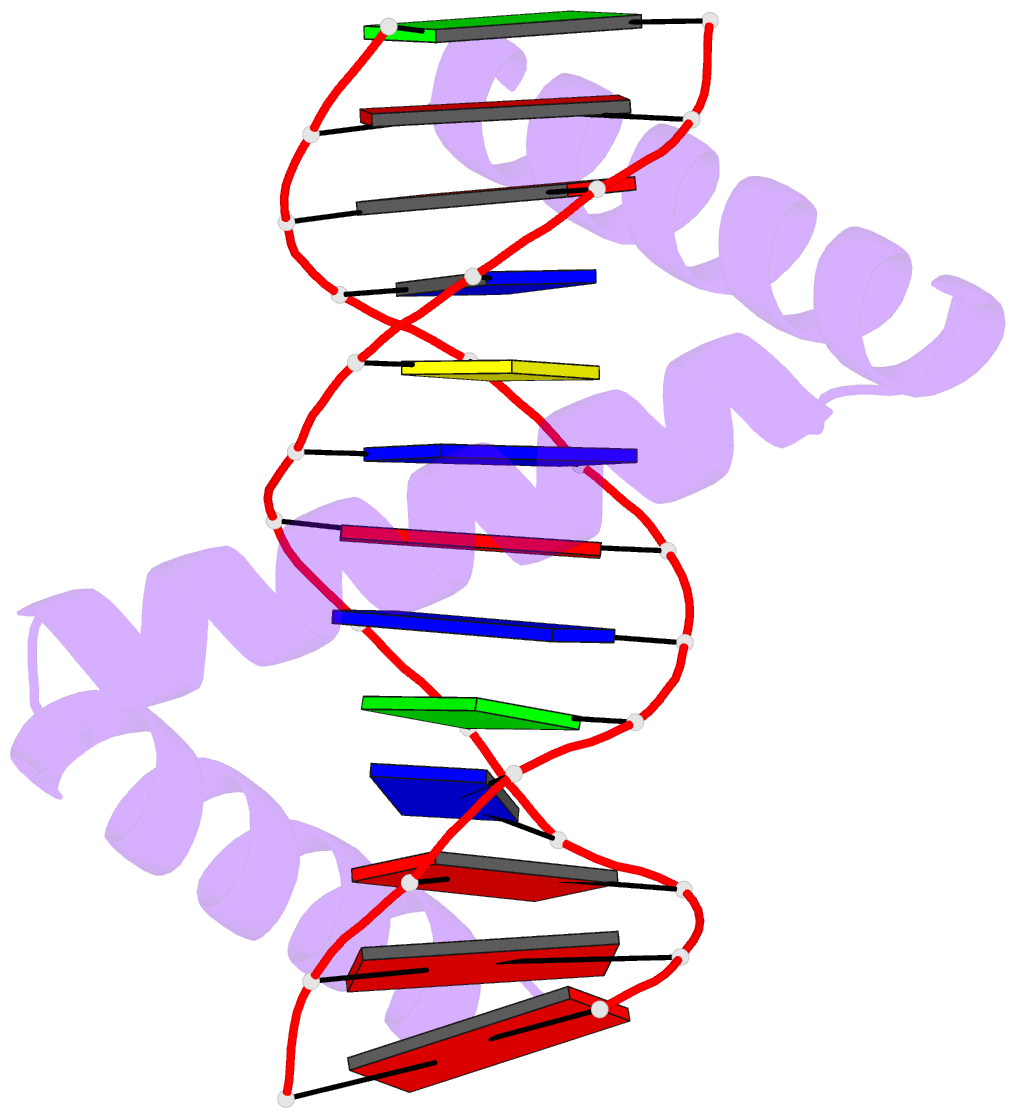
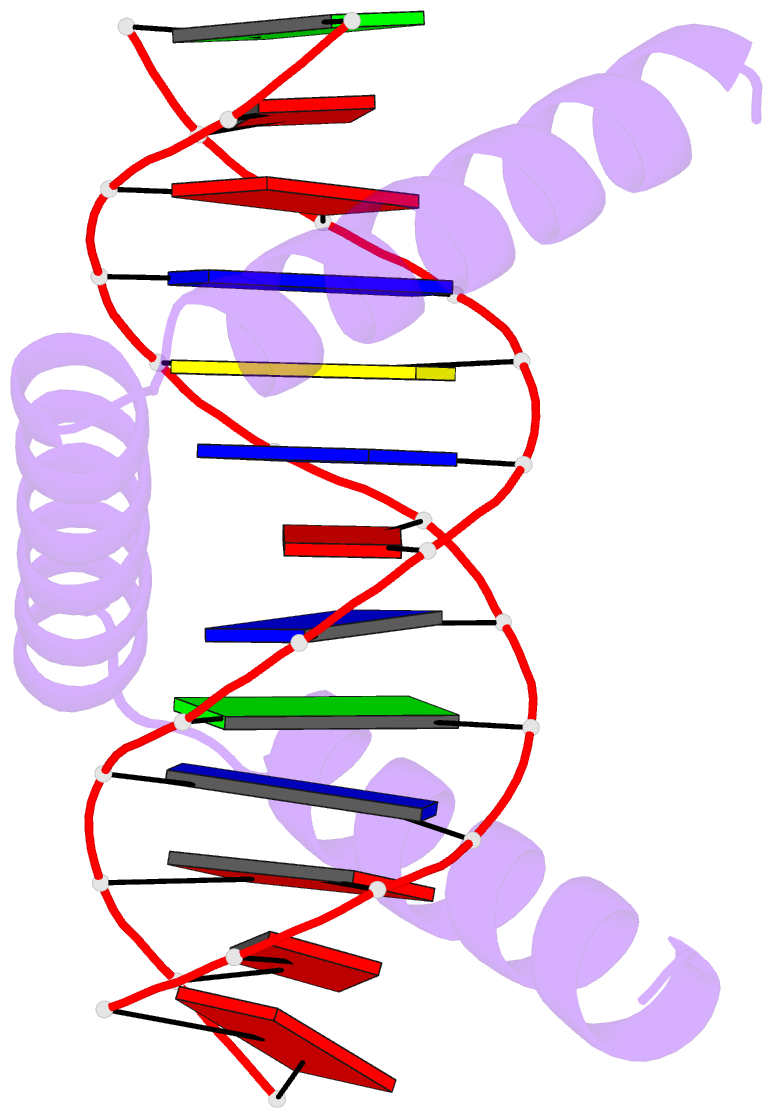
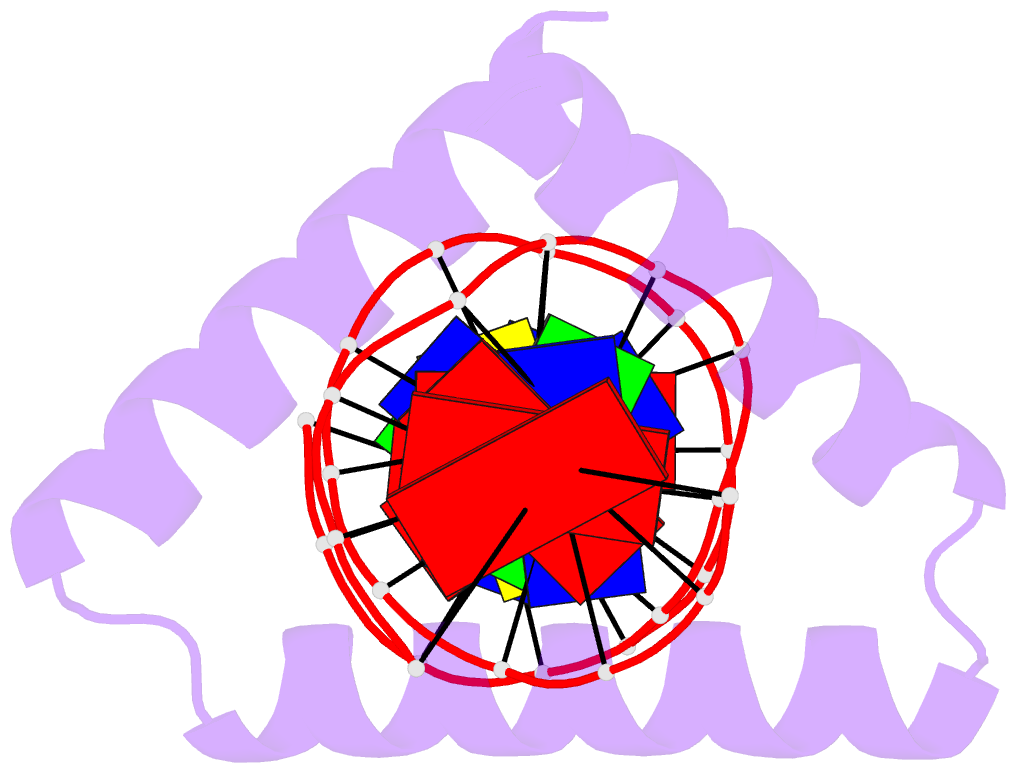
- The crystal structure of stpr from bombyx mori in complex with 13-bp DNA derived from the +290 site of fibroin gene
- Reference
- Yu LY, Cheng W, Zhou K, Li WF, Yu HM, Gao X, Shen X, Wu Q, Chen Y, Zhou CZ (2016): "Structures of an all-alpha protein running along the DNA major groove." Nucleic Acids Res. doi: 10.1093/nar/gkw133.
- Abstract
- Despite over 3300 protein-DNA complex structures have been reported in the past decades, there remain some unknown recognition patterns between protein and target DNA. The silkgland-specific transcription factor FMBP-1 from the silkworm Bombyx mori contains a unique DNA-binding domain of four tandem STPRs, namely the score and three amino acid peptide repeats. Here we report three structures of this STPR domain (termed BmSTPR) in complex with DNA of various lengths. In the presence of target DNA, BmSTPR adopts a zig-zag structure of three or four tandem α-helices that run along the major groove of DNA. Structural analyses combined with binding assays indicate BmSTPR prefers the AT-rich sequences, with each α-helix covering a DNA sequence of 4 bp. The successive AT-rich DNAs adopt a wider major groove, which is in complementary in shape and size to the tandem α-helices of BmSTPR. Substitutions of DNA sequences and affinity comparison further prove that BmSTPR recognizes the major groove mainly via shape readout. Multiple-sequence alignment suggests this unique DNA-binding pattern should be highly conserved for the STPR domain containing proteins which are widespread in animals. Together, our findings provide structural insights into the specific interactions between a novel DNA-binding protein and a unique deformed B-DNA.