Summary information and primary citation
- PDB-id
- 5dcv; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- RNA binding protein-RNA
- Method
- X-ray (3.401 Å)
- Summary
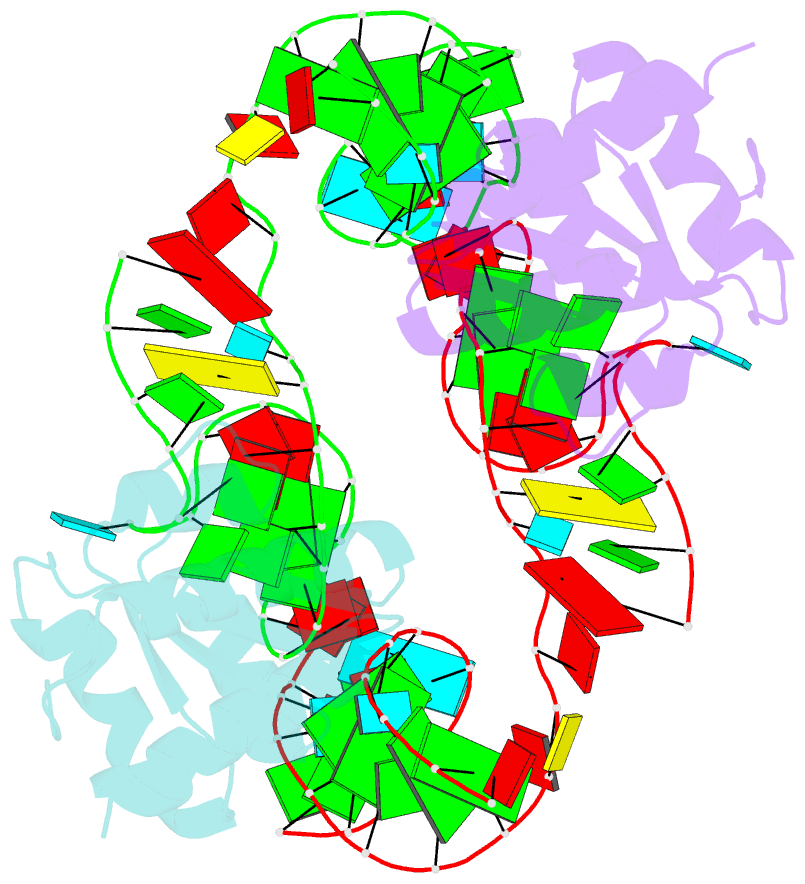
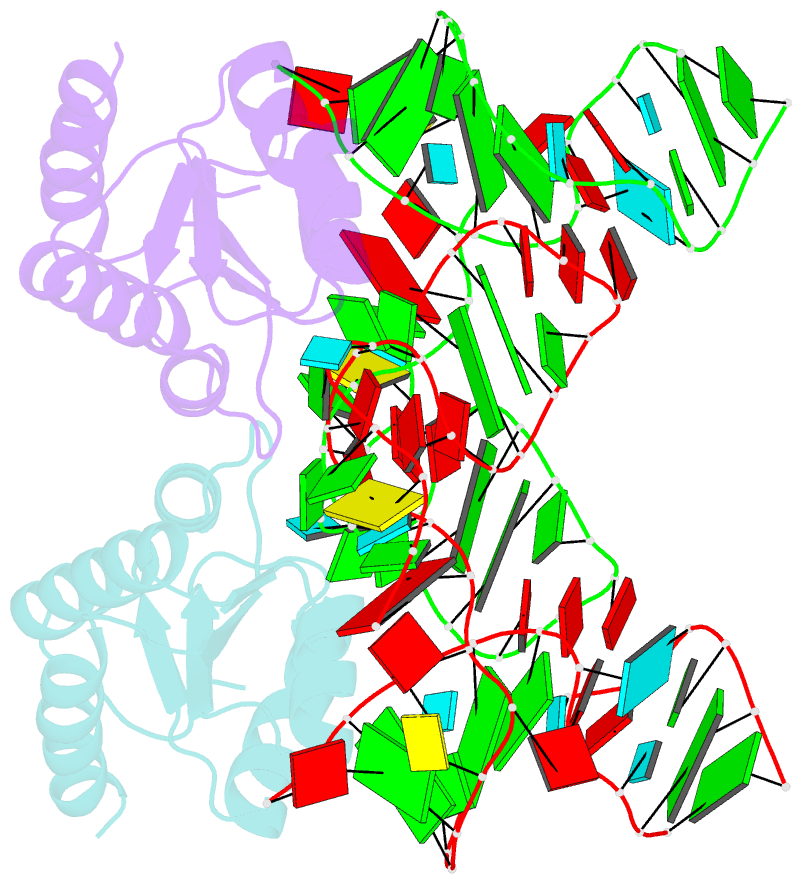
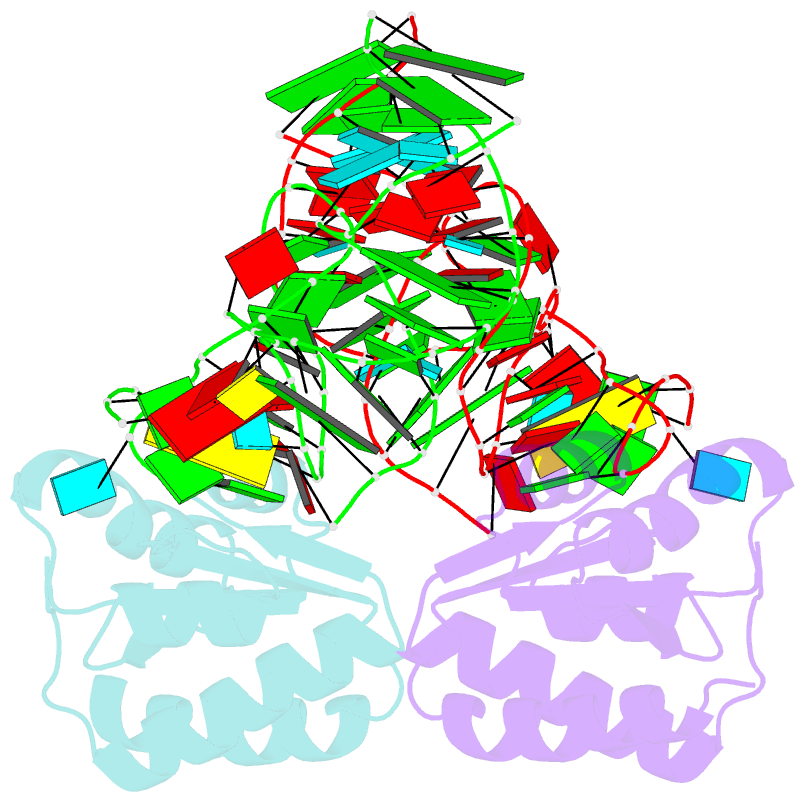
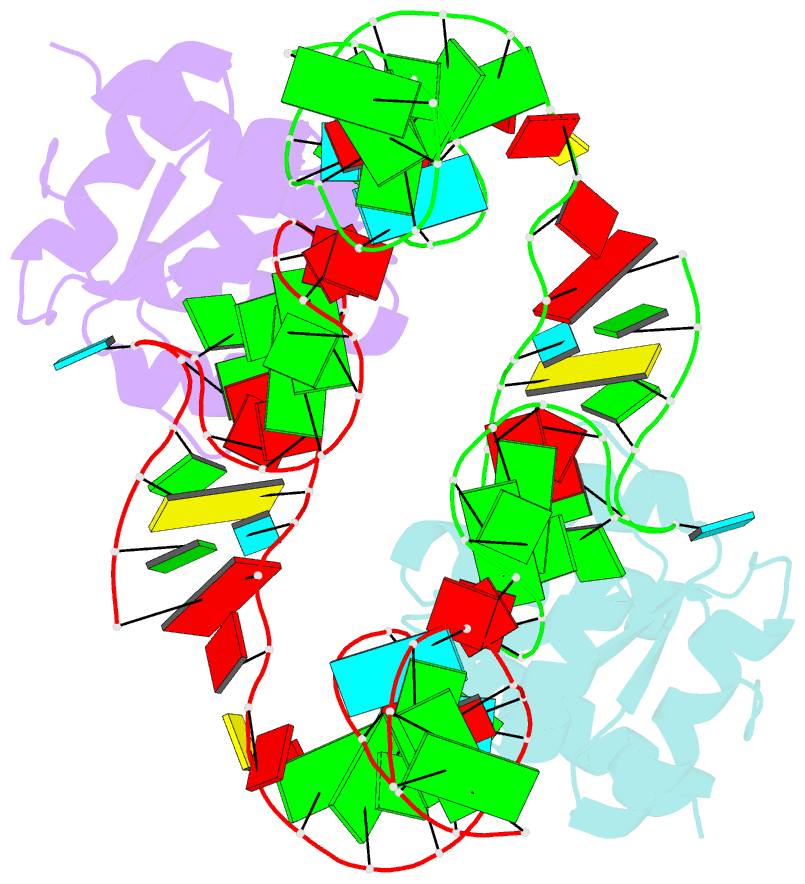
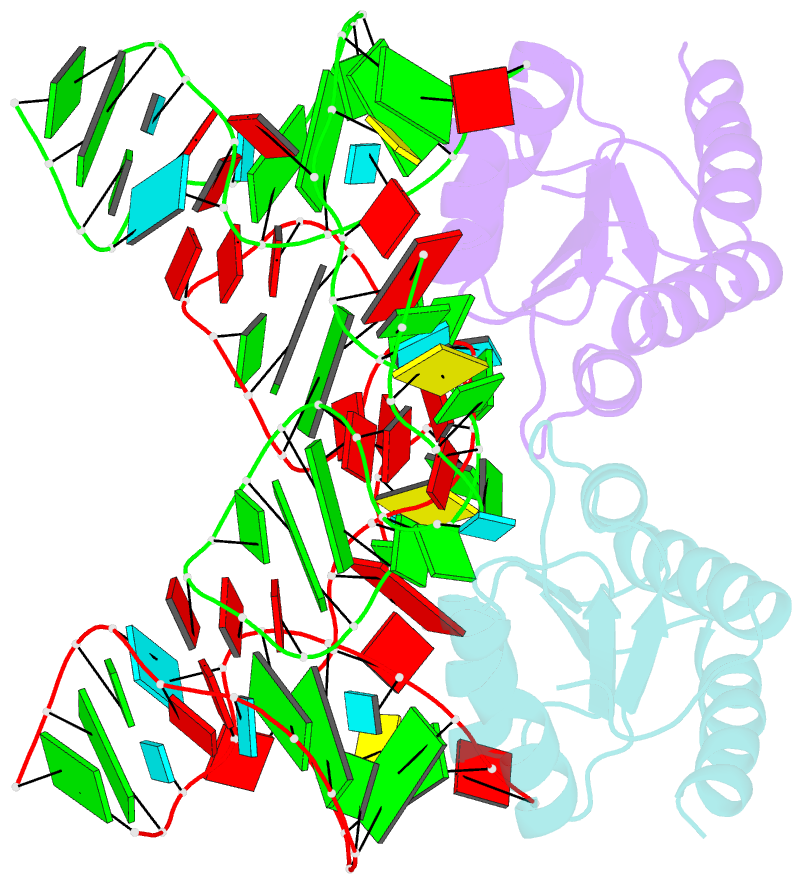
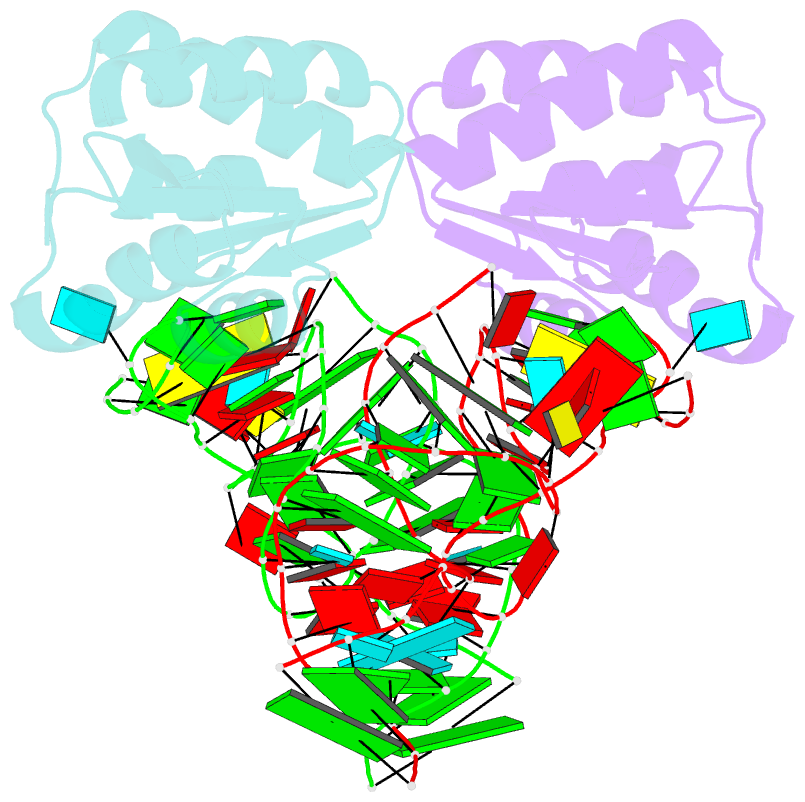
- Crystal structure of phorpp38-sl12m complex
- Reference
- Oshima K, Kakiuchi Y, Tanaka Y, Ueda T, Nakashima T, Kimura M, Yao M (2016): "Structural basis for recognition of a kink-turn motif by an archaeal homologue of human RNase P protein Rpp38." Biochem.Biophys.Res.Commun., 474, 541-546. doi: 10.1016/j.bbrc.2016.04.118.
- Abstract
- PhoRpp38 in the hyperthermophilic archaeon Pyrococcus horikoshii, a homologue of human ribonuclease P (RNase P) protein Rpp38, belongs to the ribosomal protein L7Ae family that specifically recognizes a kink-turn (K-turn) motif. A previous biochemical study showed that PhoRpp38 specifically binds to two stem-loops, SL12 and SL16, containing helices P12.1/12.2 and P15/16 respectively, in P. horikoshii RNase P RNA (PhopRNA). In order to gain insight into the PhoRpp38 binding mode to PhopRNA, we determined the crystal structure of PhoRpp38 in complex with the SL12 mutant (SL12M) at a resolution of 3.4 Å. The structure revealed that Lys35 on the β-strand (β1) and Asn38, Glu39, and Lys42 on the α-helix (α2) in PhoRpp38 interact with characteristic G•A and A•G pairs in SL12M, where Ile93, Glu94, and Val95, on a loop between α4 and β4 in PhoRpp38, interact with the 3-nucleotide bulge (G-G-U) in the SL12M. The structure demonstrates the previously proposed secondary structure of SL12, including helix P12.2. Structure-based mutational analysis indicated that amino acid residues involved in the binding to SL12 are also responsible for the binding to SL16. This result suggested that each PhoRpp38 binds to the K-turns in SL12 and SL16 in PhopRNA. A pull-down assay further suggested the presence of a second K-turn in SL12. Based on the present results, together with available data, we discuss a structural basis for recognition of K-turn motifs in PhopRNA by PhoRpp38.