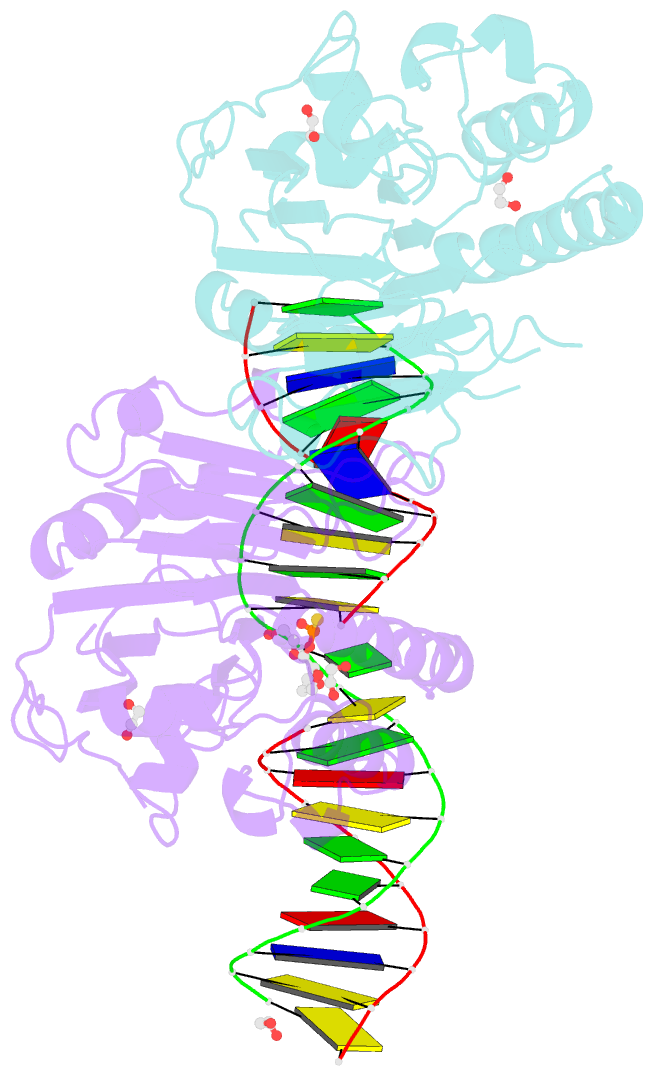
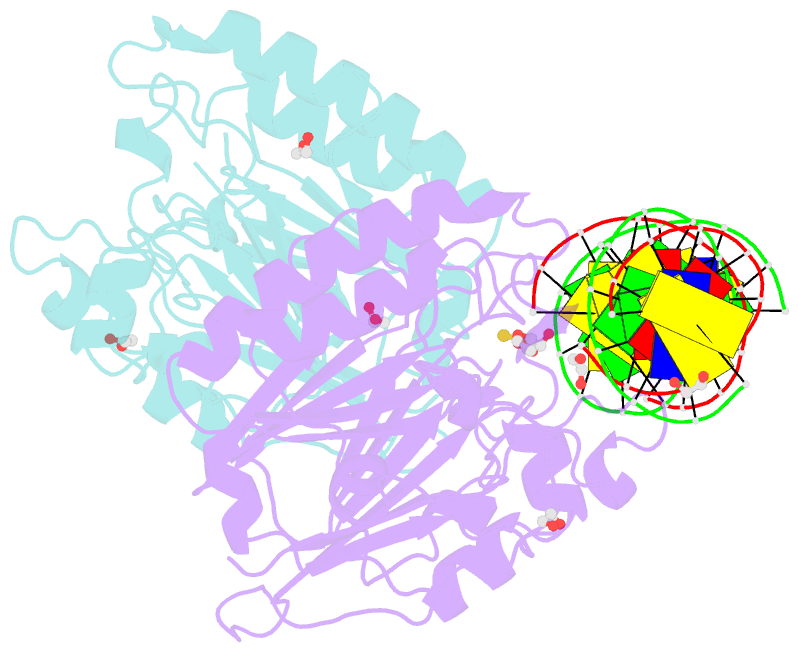
Summary information and primary citation
- PDB-id
- 5dfi; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- lyase-DNA
- Method
- X-ray (1.63 Å)
- Summary
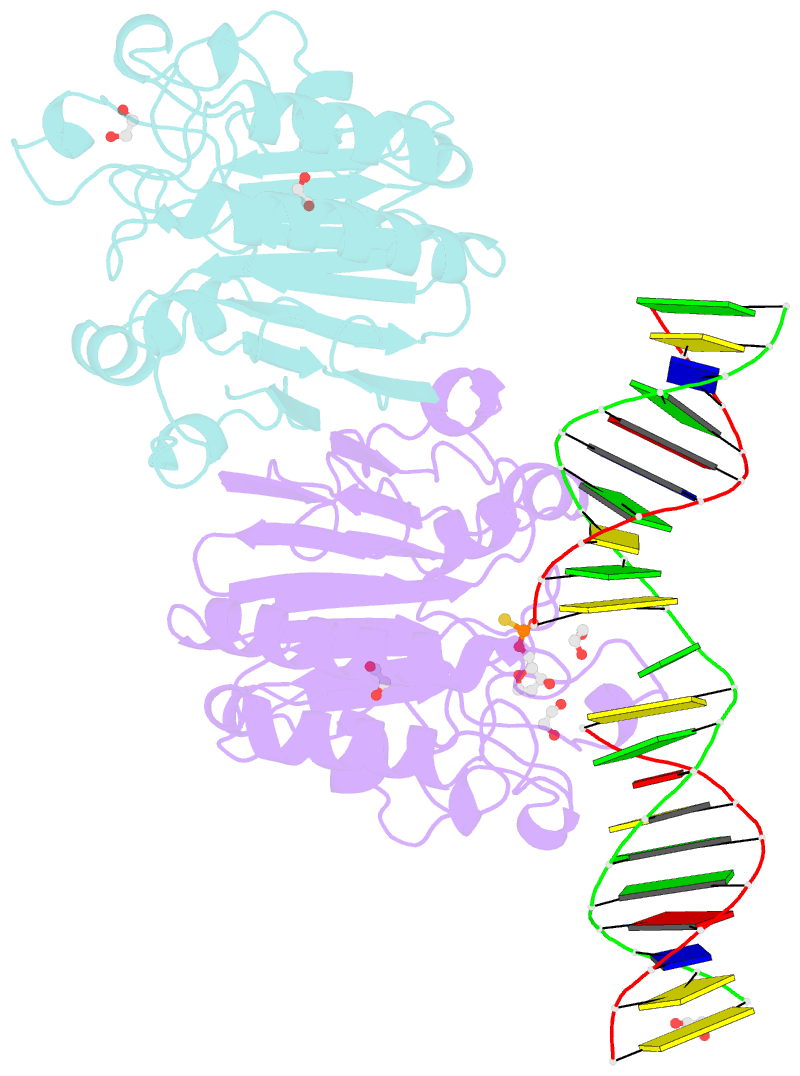
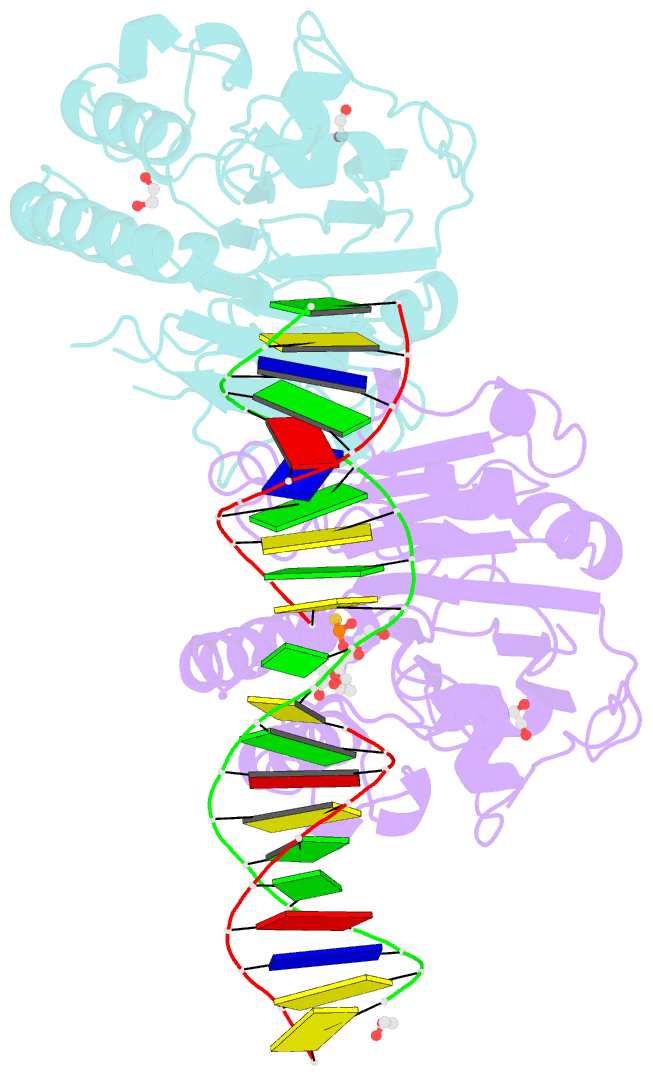
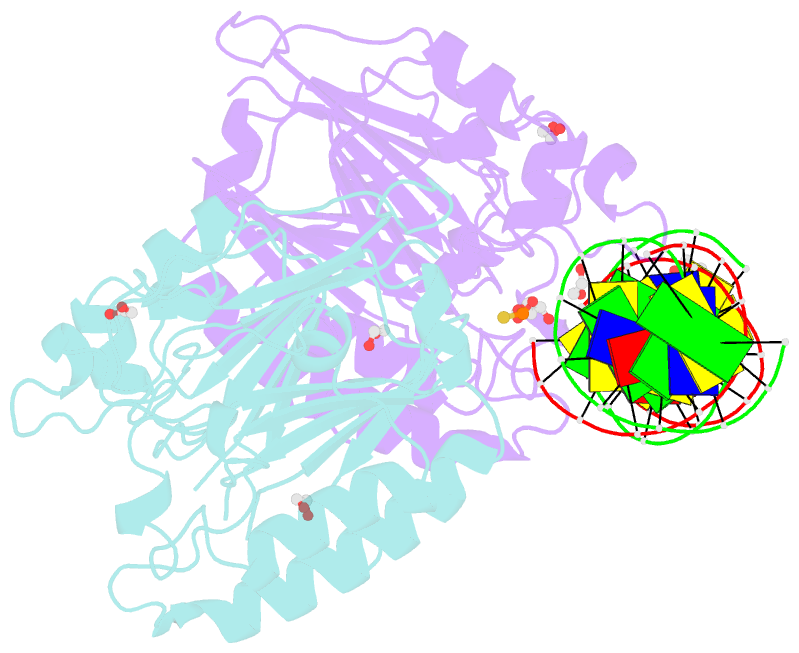
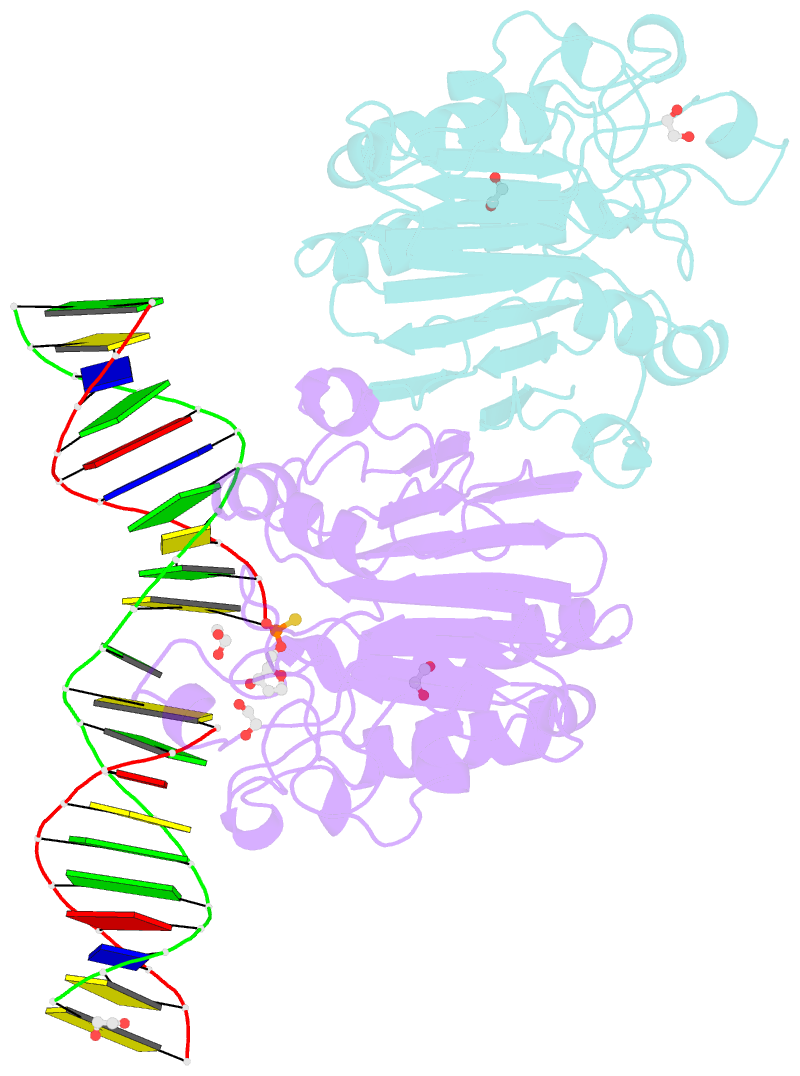
- Human ape1 phosphorothioate substrate complex
- Reference
- Freudenthal BD, Beard WA, Cuneo MJ, Dyrkheeva NS, Wilson SH (2015): "Capturing snapshots of APE1 processing DNA damage." Nat.Struct.Mol.Biol., 22, 924-931. doi: 10.1038/nsmb.3105.
- Abstract
- DNA apurinic-apyrimidinic (AP) sites are prevalent noncoding threats to genomic stability and are processed by AP endonuclease 1 (APE1). APE1 incises the AP-site phosphodiester backbone, generating a DNA-repair intermediate that is potentially cytotoxic. The molecular events of the incision reaction remain elusive, owing in part to limited structural information. We report multiple high-resolution human APE1-DNA structures that divulge new features of the APE1 reaction, including the metal-binding site, the nucleophile and the arginine clamps that mediate product release. We also report APE1-DNA structures with a T-G mismatch 5' to the AP site, representing a clustered lesion occurring in methylated CpG dinucleotides. These structures reveal that APE1 molds the T-G mismatch into a unique Watson-Crick-like geometry that distorts the active site, thus reducing incision. These snapshots provide mechanistic clarity for APE1 while affording a rational framework to manipulate biological responses to DNA damage.