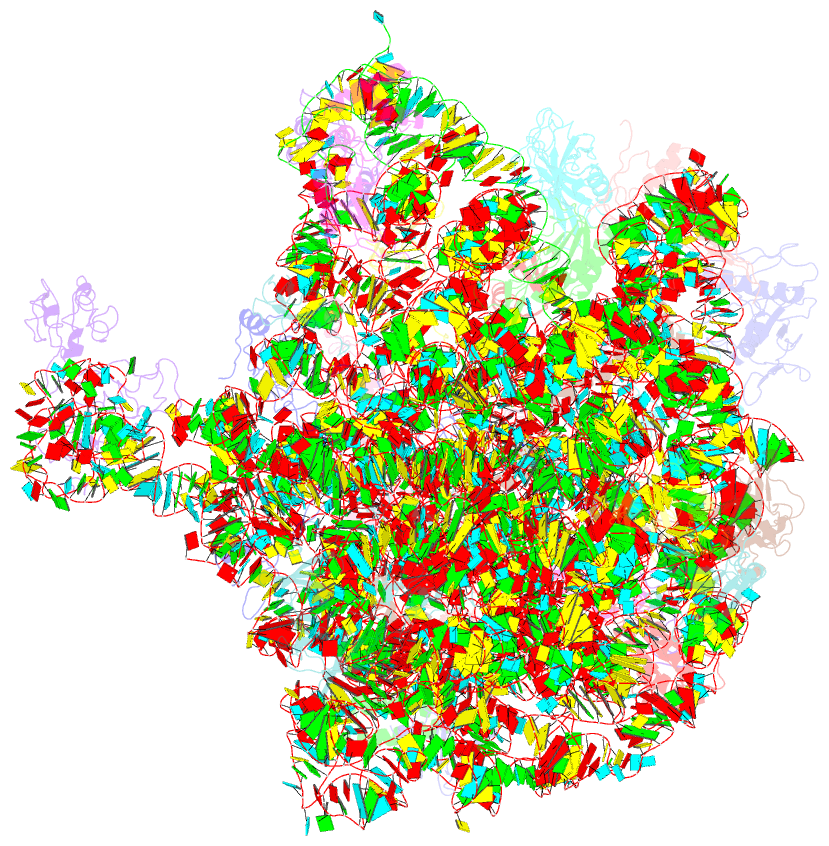
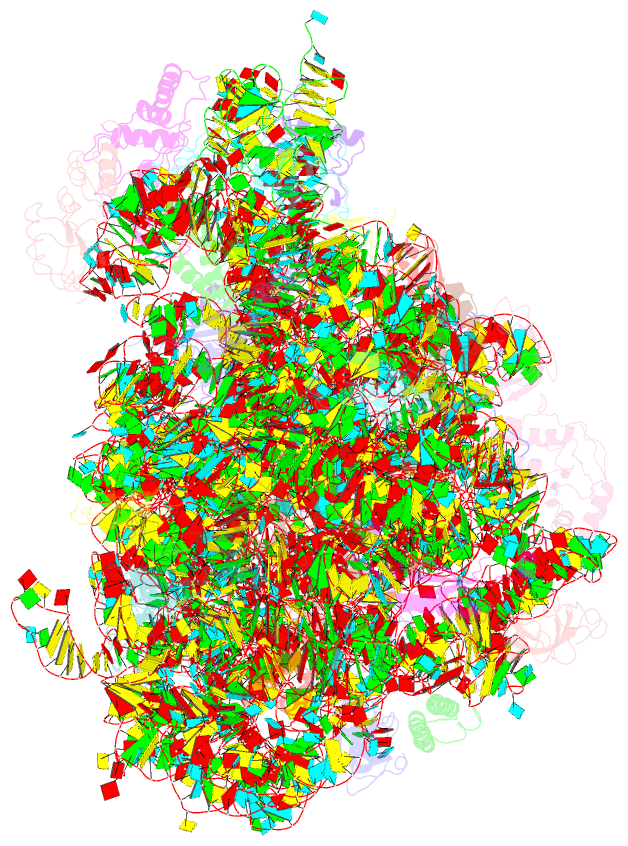
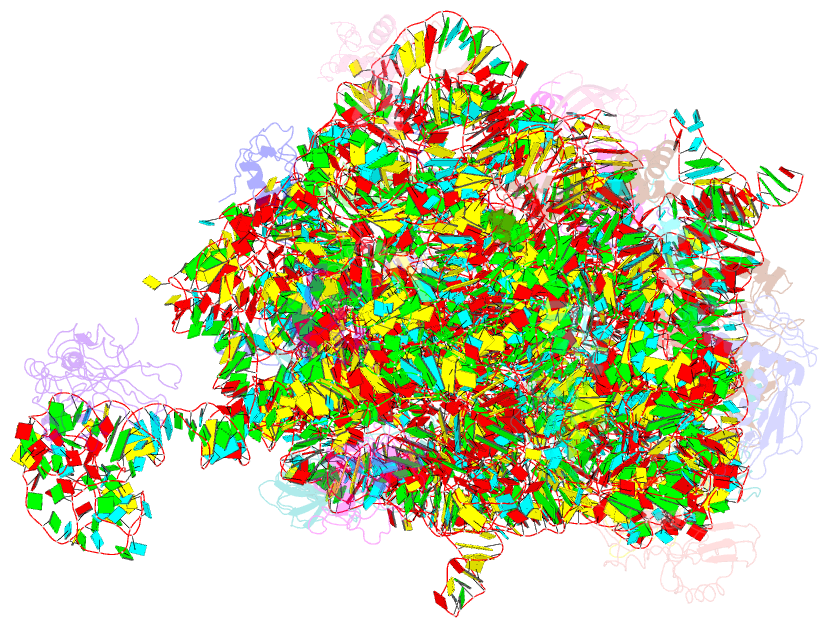
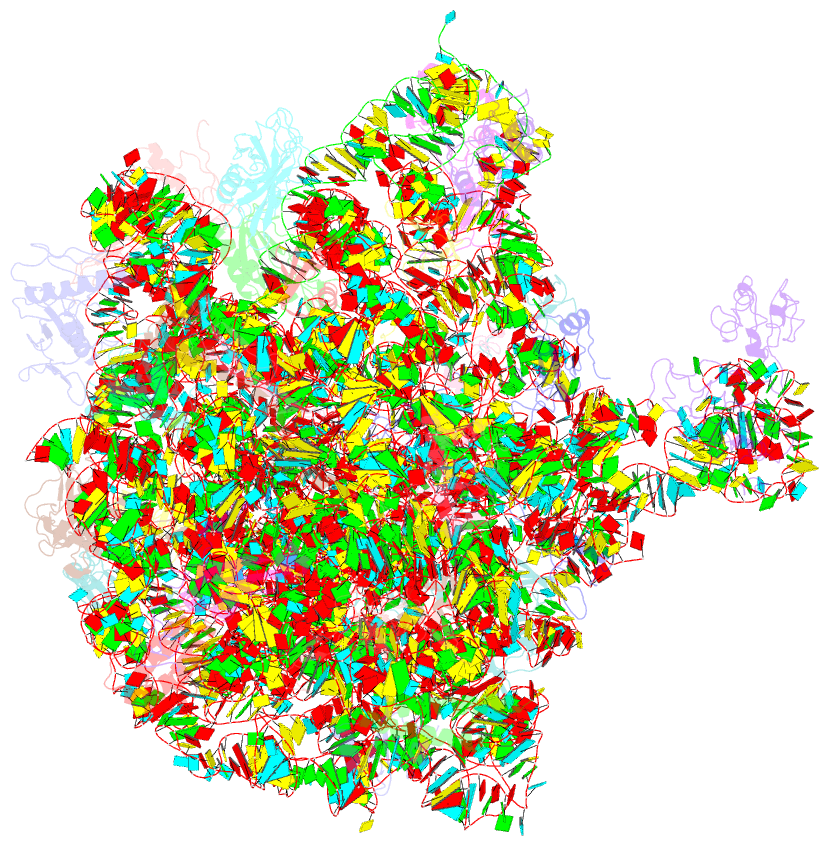
Summary information and primary citation
- PDB-id
- 5dm7; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- ribosome
- Method
- X-ray (3.0 Å)
- Summary
- Crystal structure of the 50s ribosomal subunit from deinococcus radiodurans in complex with hygromycin a
- Reference
- Kaminishi T, Schedlbauer A, Fabbretti A, Brandi L, Ochoa-Lizarralde B, He CG, Milon P, Connell SR, Gualerzi CO, Fucini P (2015): "Crystallographic characterization of the ribosomal binding site and molecular mechanism of action of Hygromycin A." Nucleic Acids Res., 43, 10015-10025. doi: 10.1093/nar/gkv975.
- Abstract
- Hygromycin A (HygA) binds to the large ribosomal subunit and inhibits its peptidyl transferase (PT) activity. The presented structural and biochemical data indicate that HygA does not interfere with the initial binding of aminoacyl-tRNA to the A site, but prevents its subsequent adjustment such that it fails to act as a substrate in the PT reaction. Structurally we demonstrate that HygA binds within the peptidyl transferase center (PTC) and induces a unique conformation. Specifically in its ribosomal binding site HygA would overlap and clash with aminoacyl-A76 ribose moiety and, therefore, its primary mode of action involves sterically restricting access of the incoming aminoacyl-tRNA to the PTC.