Summary information and primary citation
- PDB-id
- 5dto; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- viral protein-RNA
- Method
- X-ray (2.603 Å)
- Summary
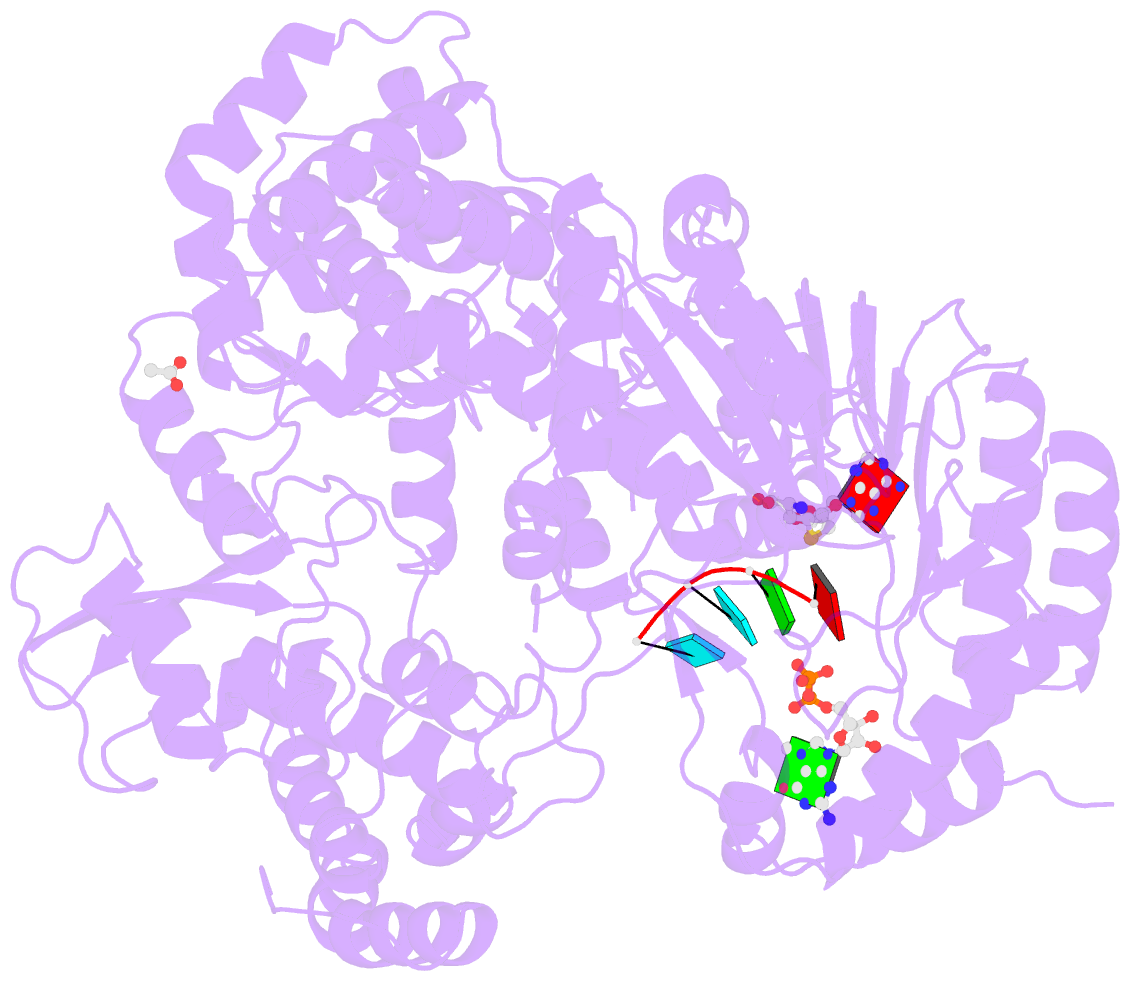
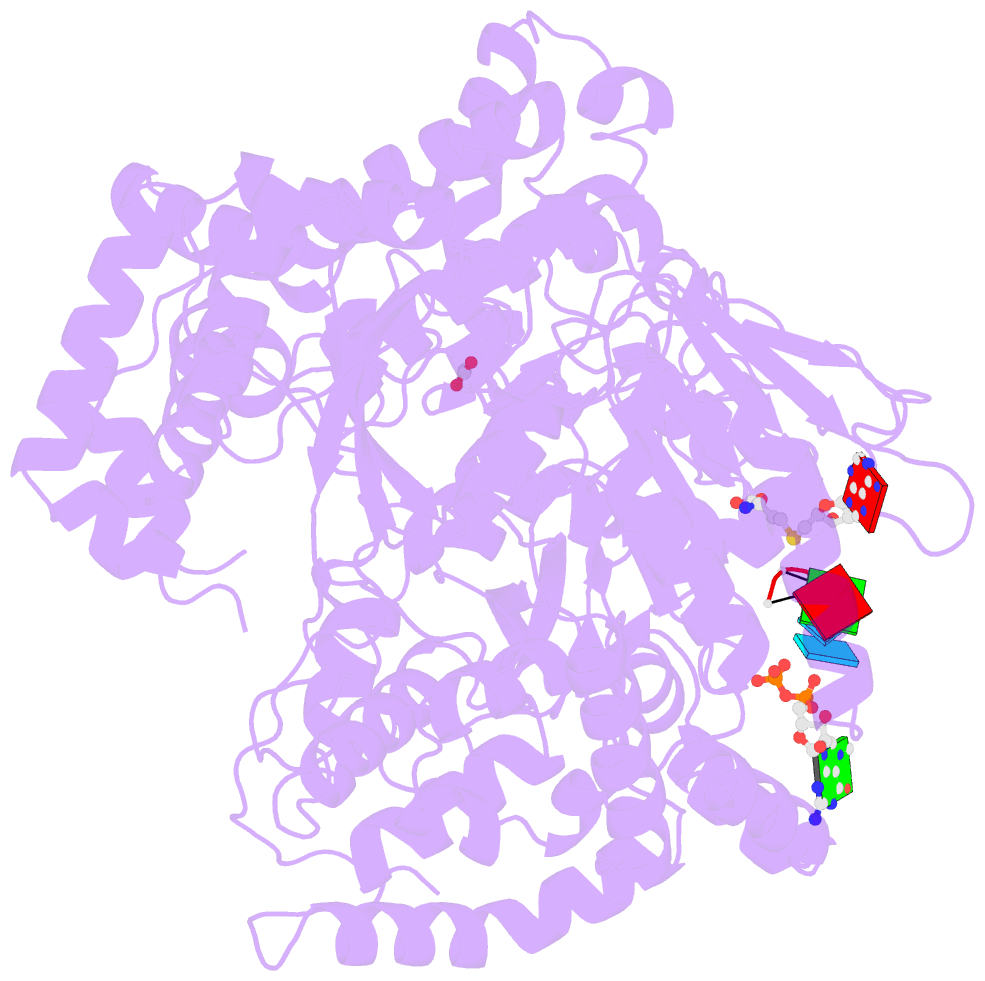
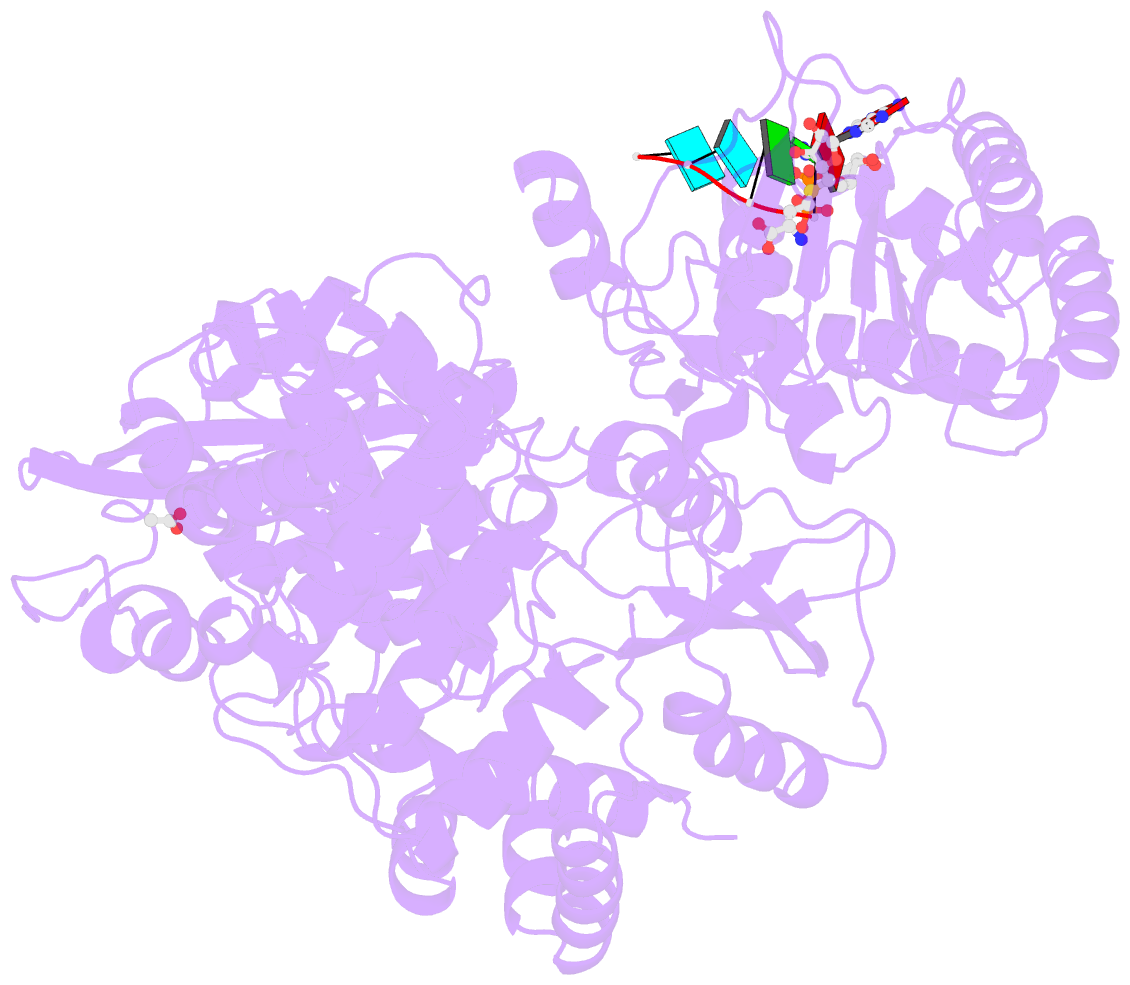
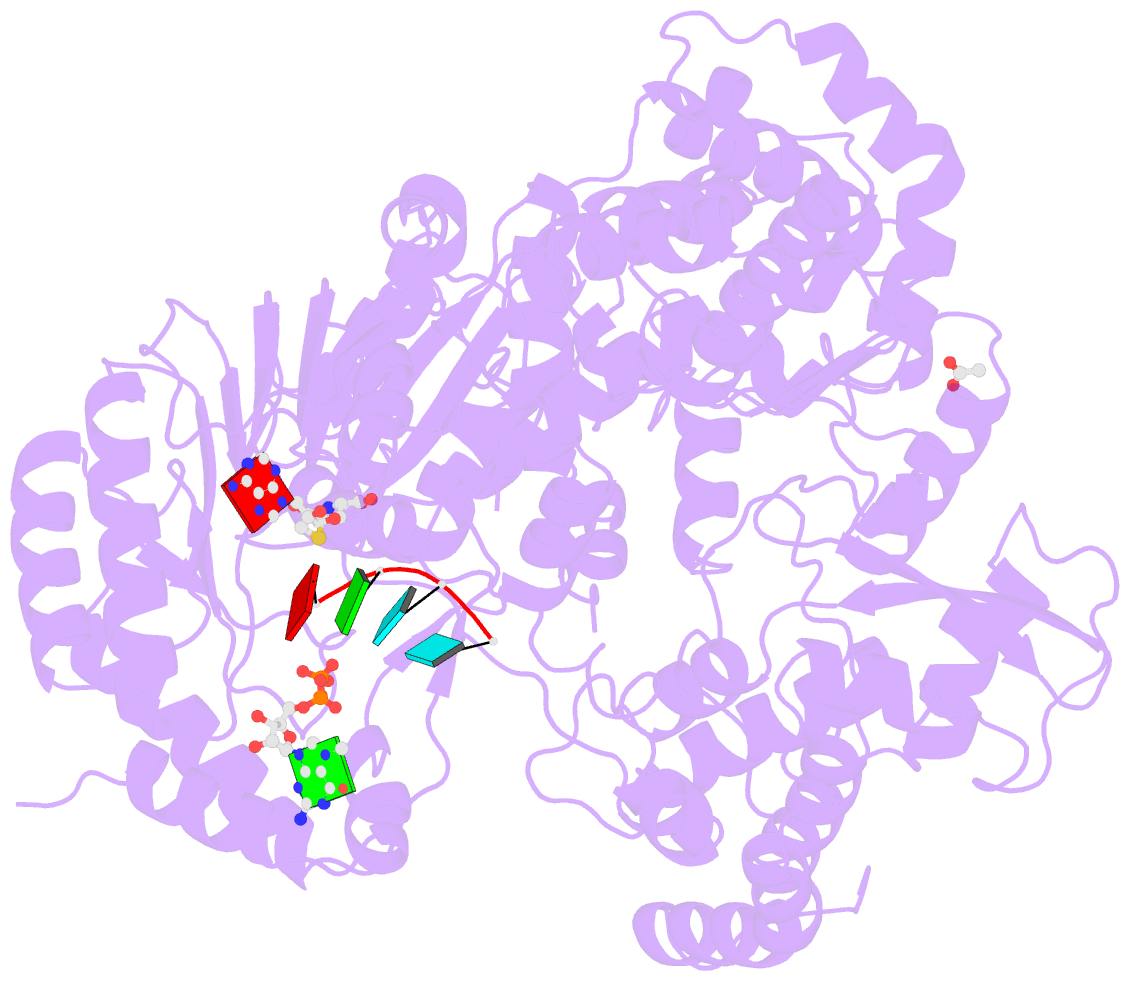
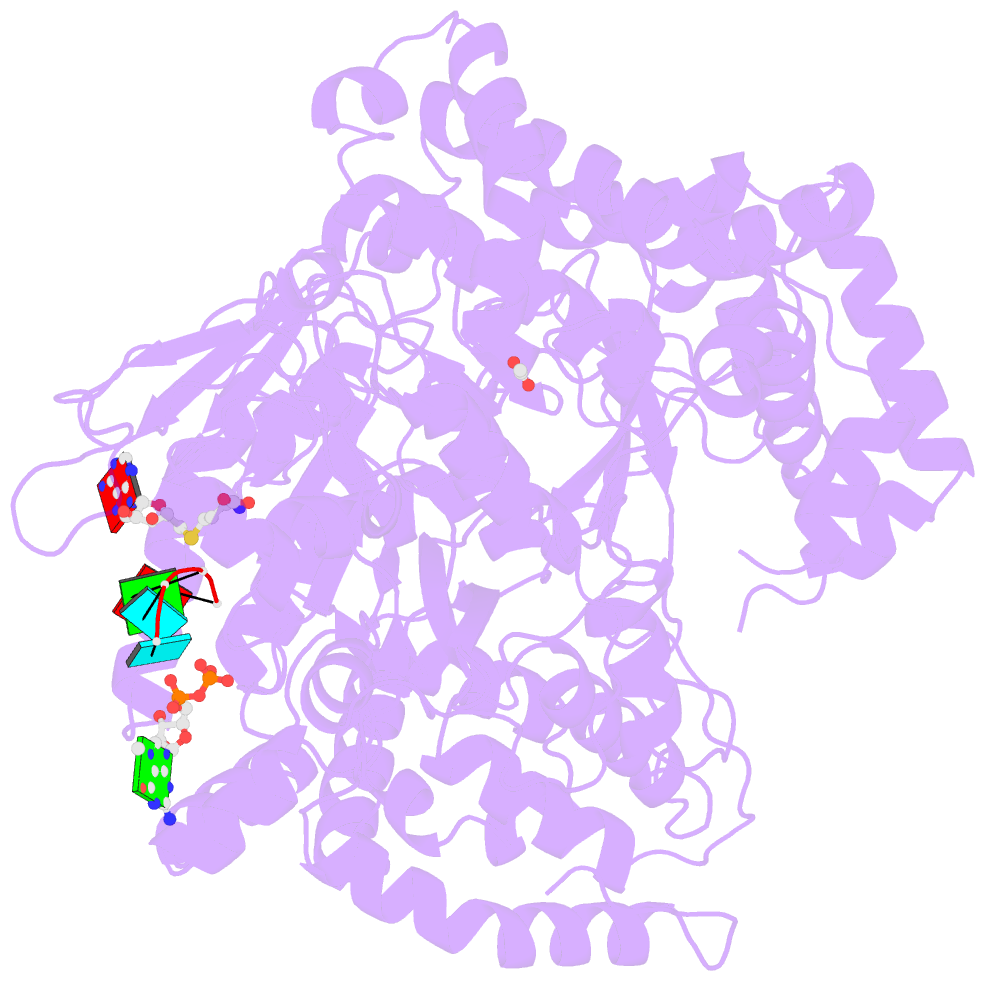
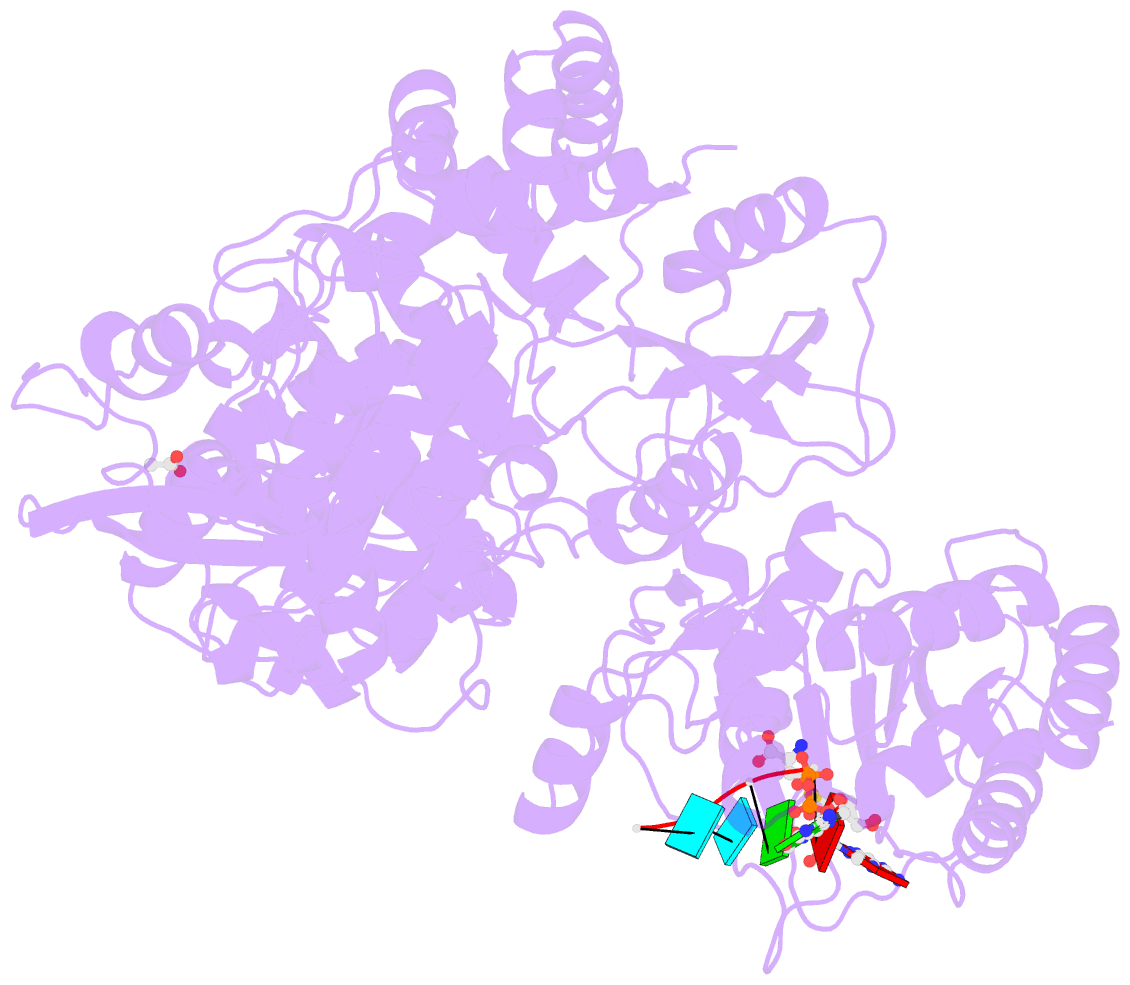
- Dengue virus full length ns5 complexed with viral cap 0-RNA and sah
- Reference
- Zhao Y, Soh TS, Lim SP, Chung KY, Swaminathan K, Vasudevan SG, Shi P-Y, Lescar J, Luo D (2015): "Molecular basis for specific viral RNA recognition and 2'-O-ribose methylation by the dengue virus nonstructural protein 5 (NS5)." Proc.Natl.Acad.Sci.USA, 112, 14834-14839. doi: 10.1073/pnas.1514978112.
- Abstract
- Dengue virus (DENV) causes several hundred million human infections and more than 20,000 deaths annually. Neither an efficacious vaccine conferring immunity against all four circulating serotypes nor specific drugs are currently available to treat this emerging global disease. Capping of the DENV RNA genome is an essential structural modification that protects the RNA from degradation by 5' exoribonucleases, ensures efficient expression of viral proteins, and allows escape from the host innate immune response. The large flavivirus nonstructural protein 5 (NS5) (105 kDa) has RNA methyltransferase activities at its N-terminal region, which is responsible for capping the virus RNA genome. The methyl transfer reactions are thought to occur sequentially using the strictly conserved flavivirus 5' RNA sequence as substrate (GpppAG-RNA), leading to the formation of the 5' RNA cap: G0pppAG-RNA → (m7)G0pppAG-RNA ("cap-0")→(m7)G0pppAm2'-O-G-RNA ("cap-1"). To elucidate how viral RNA is specifically recognized and methylated, we determined the crystal structure of a ternary complex between the full-length NS5 protein from dengue virus, an octameric cap-0 viral RNA substrate bearing the authentic DENV genomic sequence (5'-(m7)G0pppA1G2U3U4G5U6U7-3'), and S-adenosyl-l-homocysteine (SAH), the by-product of the methylation reaction. The structure provides for the first time, to our knowledge, a molecular basis for specific adenosine 2'-O-methylation, rationalizes mutagenesis studies targeting the K61-D146-K180-E216 enzymatic tetrad as well as residues lining the RNA binding groove, and offers previously unidentified mechanistic and evolutionary insights into cap-1 formation by NS5, which underlies innate immunity evasion by flaviviruses.