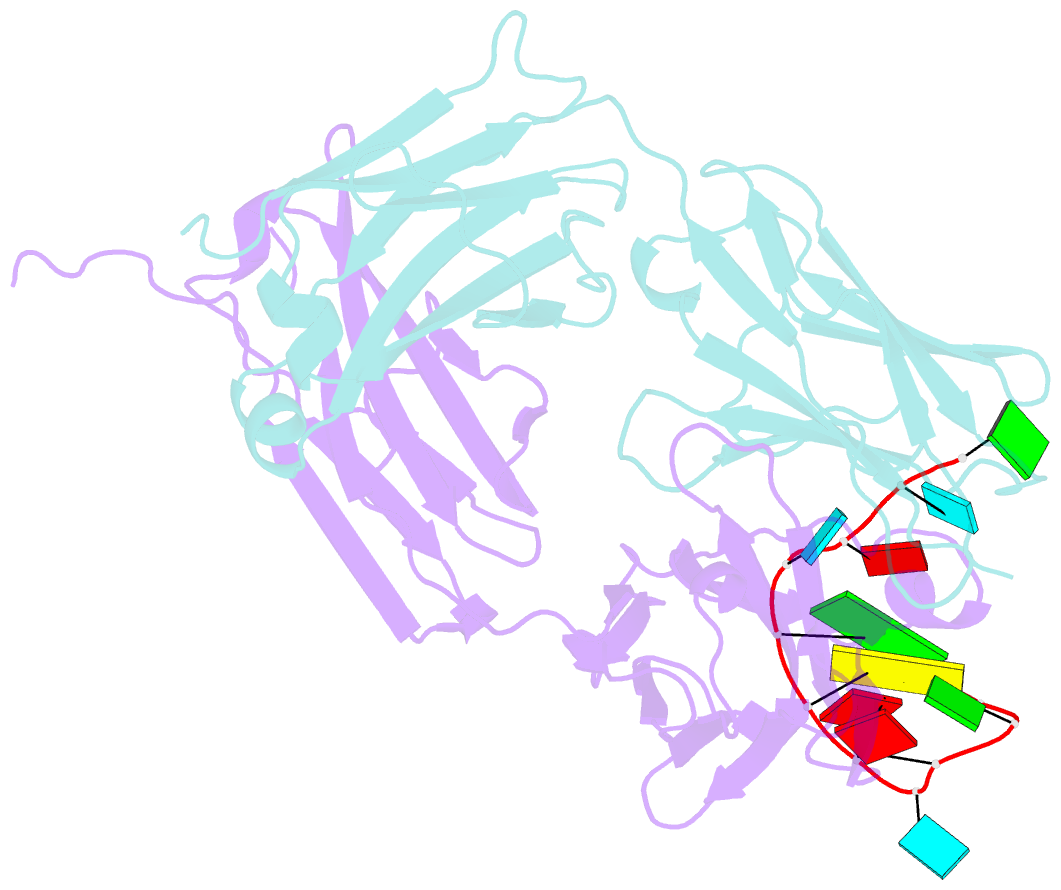
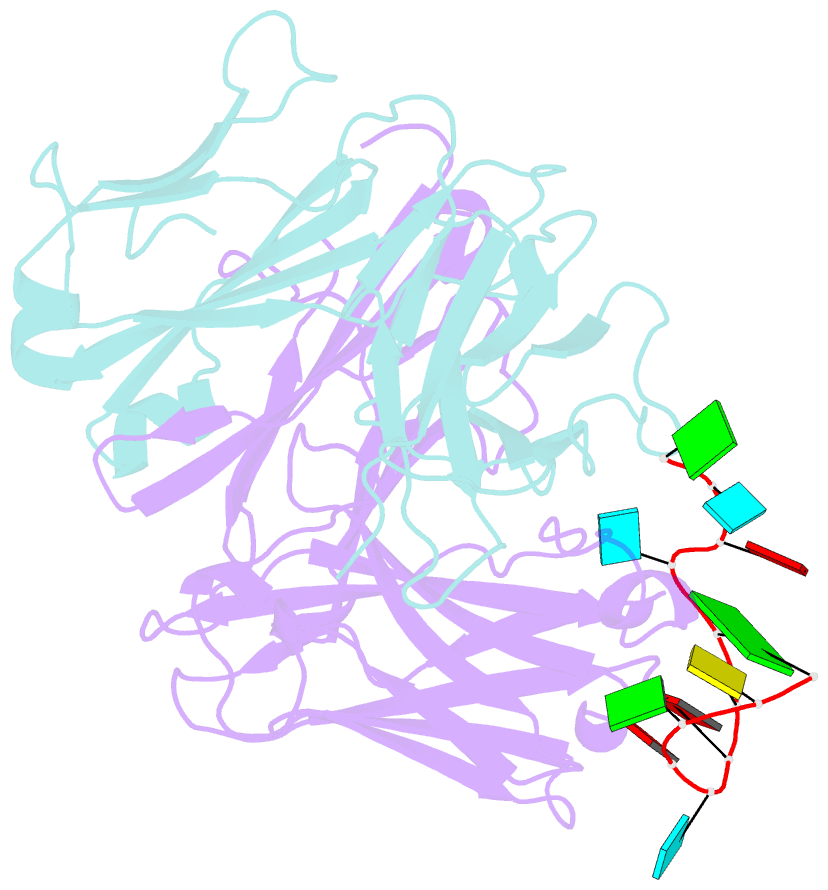
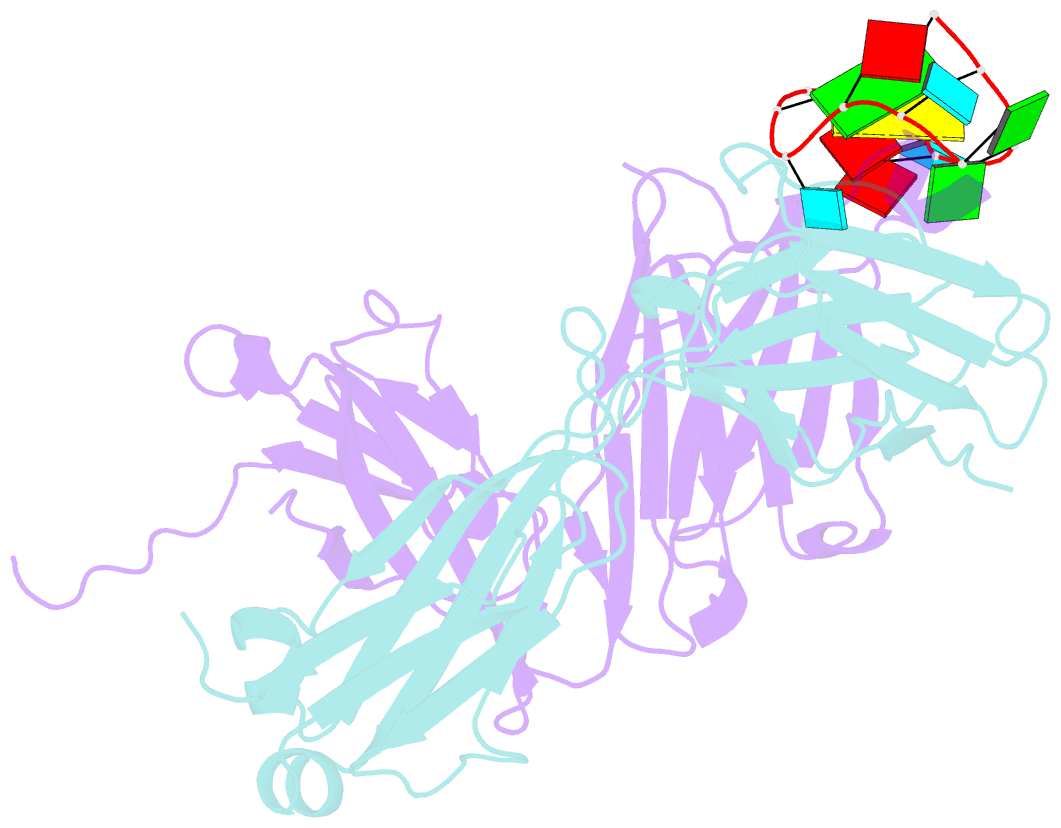
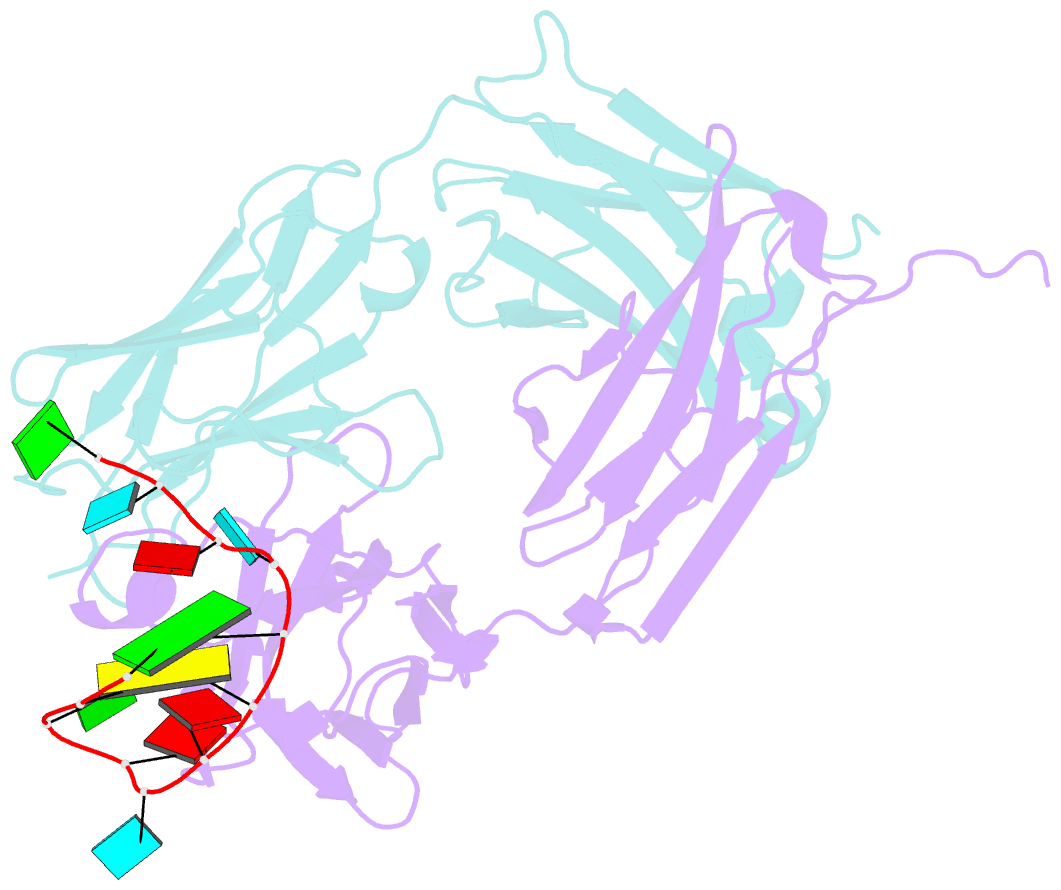
Summary information and primary citation
- PDB-id
- 5e08; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- immune system-RNA
- Method
- X-ray (2.38 Å)
- Summary
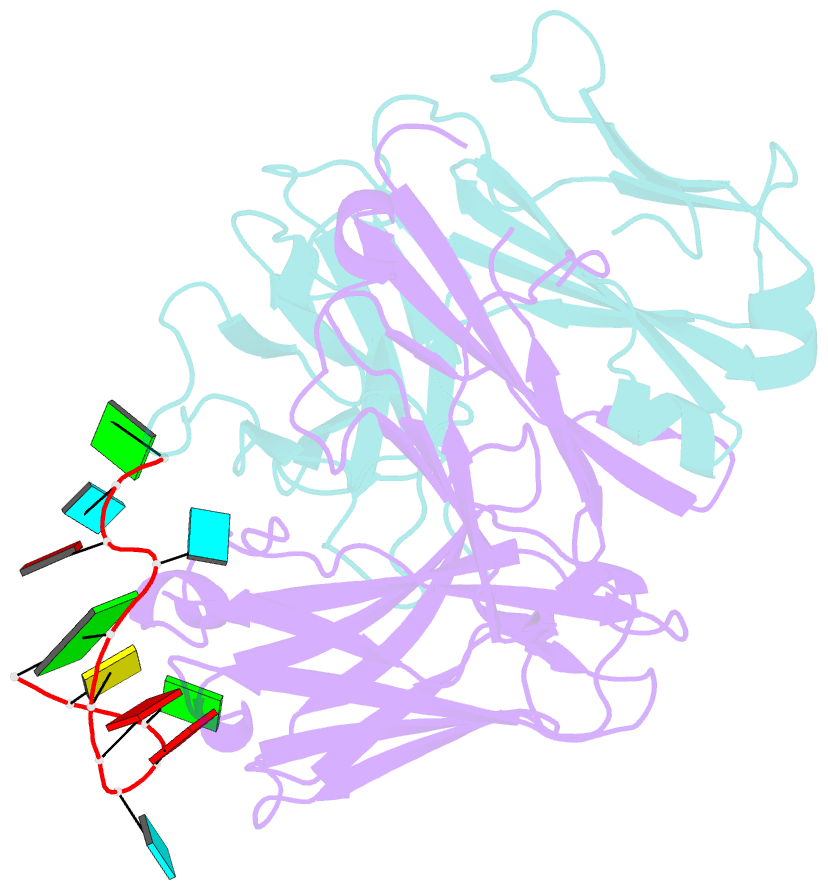
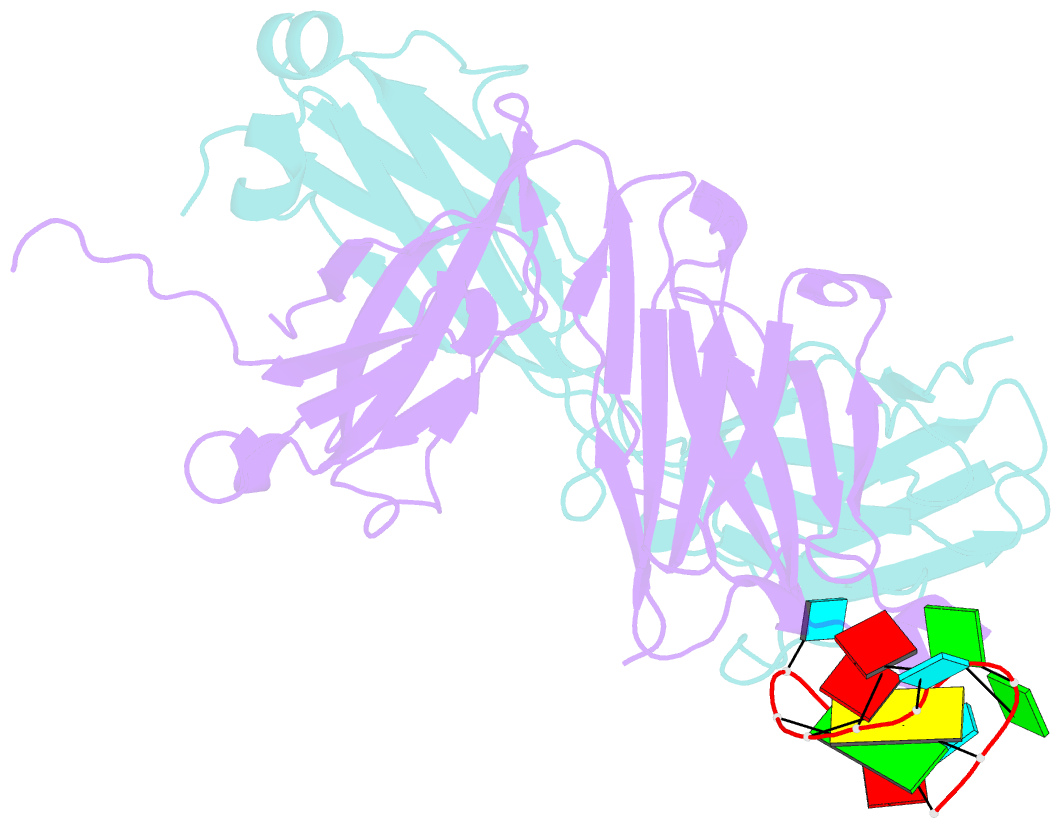
- Specific recognition of a single-stranded RNA sequence by an engineered synthetic antibody fragment
- Reference
- Shao Y, Huang H, Qin D, Li NS, Koide A, Staley JP, Koide S, Kossiakoff AA, Piccirilli JA (2016): "Specific Recognition of a Single-Stranded RNA Sequence by a Synthetic Antibody Fragment." J.Mol.Biol., 428, 4100-4114. doi: 10.1016/j.jmb.2016.08.029.
- Abstract
- Antibodies that bind RNA represent an unrealized source of reagents for synthetic biology and for characterizing cellular transcriptomes. However, facile access to RNA-binding antibodies requires the engineering of effective Fab libraries guided by the knowledge of the principles that govern RNA recognition. Here, we describe a Fab identified from a minimalist synthetic library during phage display against a branched RNA target. The Fab (BRG) binds with 20nM dissociation constant to a single-stranded RNA (ssRNA) sequence adjacent to the branch site and can block the action of debranchase enzyme. We report the crystal structure in complex with RNA target at 2.38Å. The Fab traps the RNA in a hairpin conformation that contains a 2-bp duplex capped by a tetraloop. The paratope surface consists of residues located in four complementarity-determining regions including a major contribution from H3, which adopts a helical structure that projects into a deep, wide groove formed by the RNA. The amino acid composition of the paratope reflects the library diversity, consisting mostly of tyrosine and serine residues and a small but significant contribution from a single arginine residue. This structure, involving the recognition of ssRNA via a stem-loop conformation, together with our two previous structures involving the recognition of an RNA hairpin loop and an RNA tertiary structure, reveals the capacity of minimalist libraries biased with tyrosine, serine, glycine, and arginine to form binding surfaces for specific RNA conformations and distinct levels of RNA structural hierarchy.