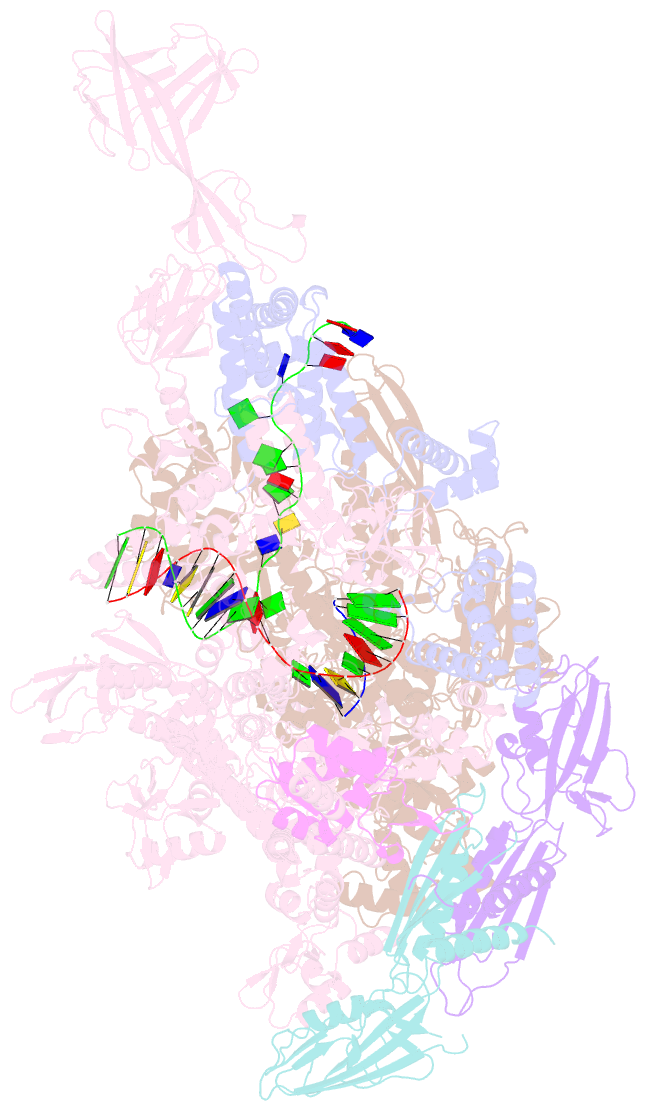
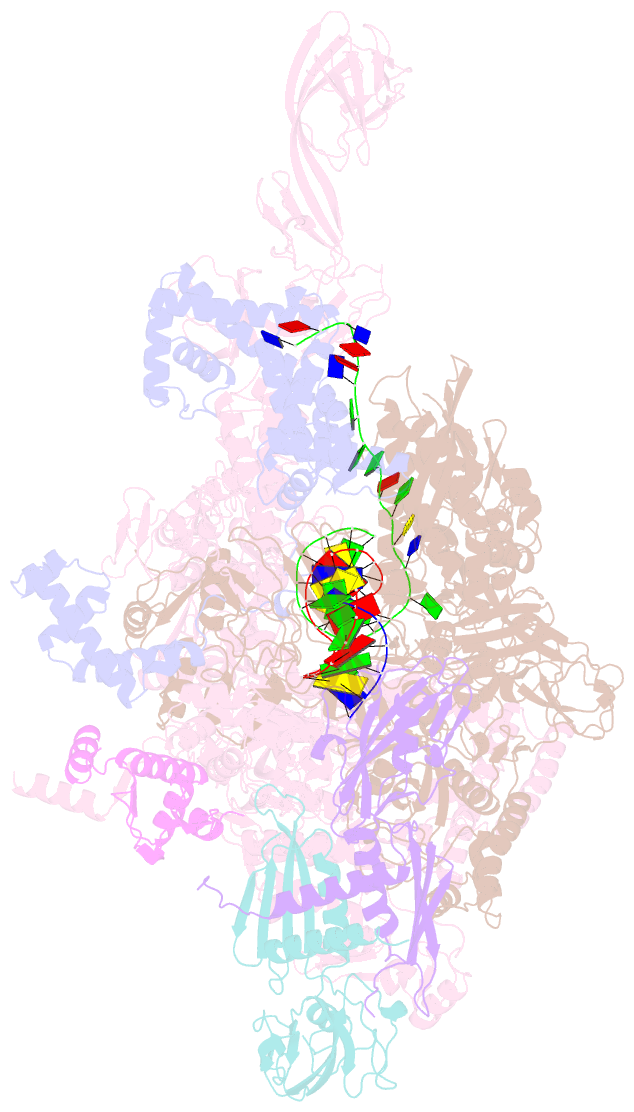
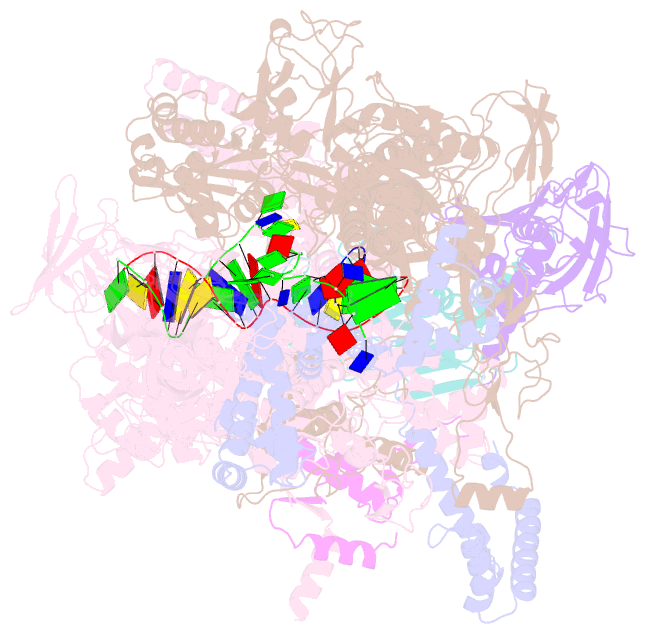
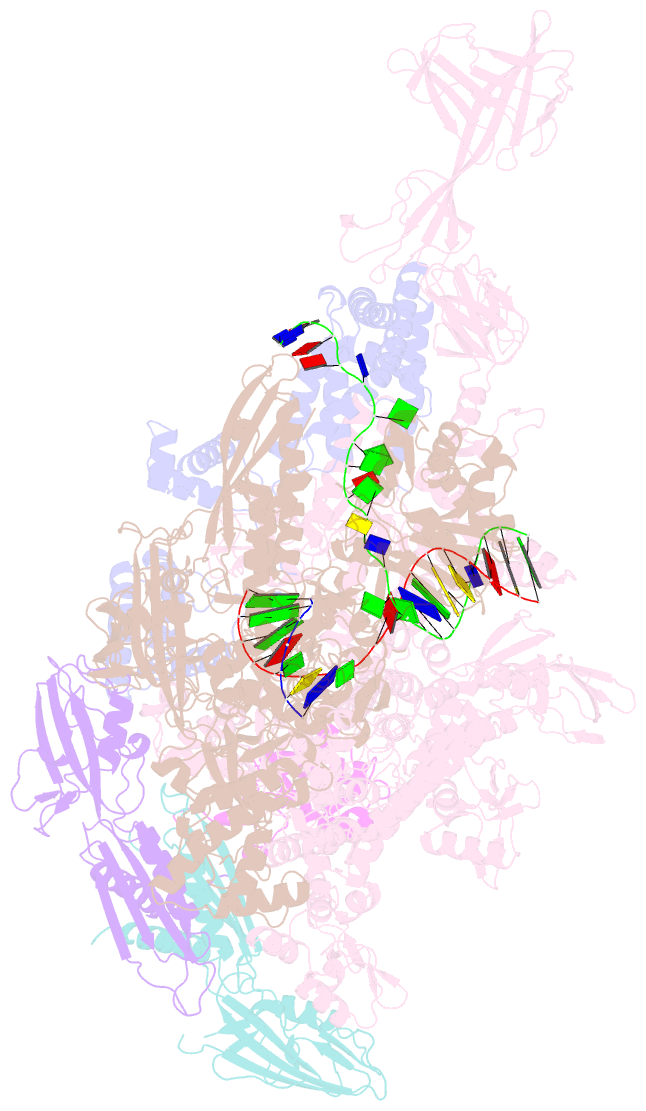
Summary information and primary citation
- PDB-id
- 5e17; SNAP-derived features in text and JSON formats;
DNAproDB
- Class
- transcription-DNA-RNA
- Method
- X-ray (3.2 Å)
- Summary
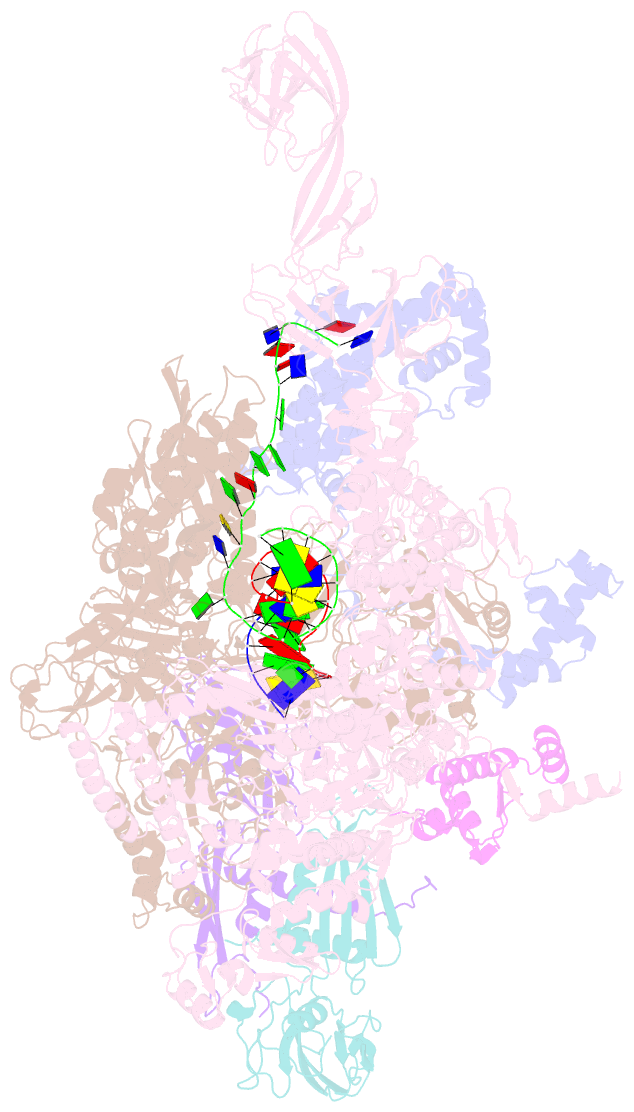
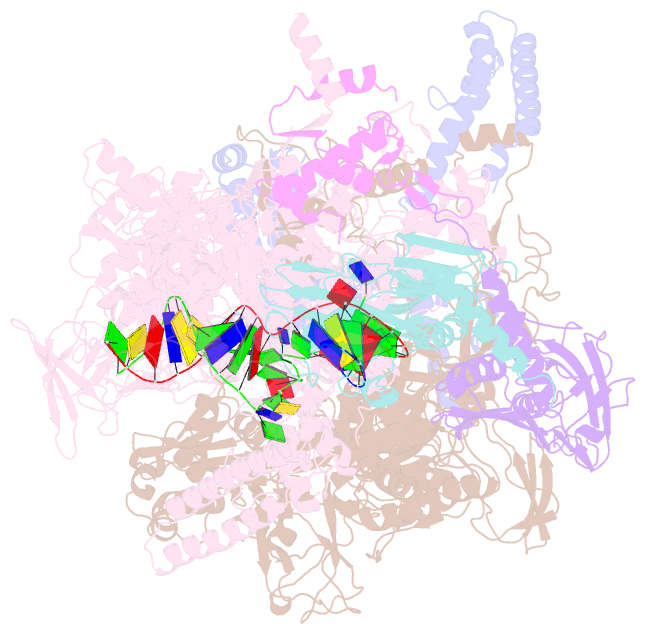
- T. thermophilus transcription initiation complex having a rrr discriminator sequence and a nontemplate-strand length corresponding to tss selection at position 7 (rpo-ggg-7)
- Reference
- Winkelman JT, Vvedenskaya IO, Zhang Y, Zhang Y, Bird JG, Taylor DM, Gourse RL, Ebright RH, Nickels BE (2016): "Multiplexed protein-DNA cross-linking: Scrunching in transcription start site selection." Science, 351, 1090-1093. doi: 10.1126/science.aad6881.
- Abstract
- In bacterial transcription initiation, RNA polymerase (RNAP) selects a transcription start site (TSS) at variable distances downstream of core promoter elements. Using next-generation sequencing and unnatural amino acid-mediated protein-DNA cross-linking, we have determined, for a library of 4(10) promoter sequences, the TSS, the RNAP leading-edge position, and the RNAP trailing-edge position. We find that a promoter element upstream of the TSS, the "discriminator," participates in TSS selection, and that, as the TSS changes, the RNAP leading-edge position changes, but the RNAP trailing-edge position does not change. Changes in the RNAP leading-edge position, but not the RNAP trailing-edge position, are a defining hallmark of the "DNA scrunching" that occurs concurrent with RNA synthesis in initial transcription. We propose that TSS selection involves DNA scrunching prior to RNA synthesis.